
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,209,403 – 18,209,510 |
| Length | 107 |
| Max. P | 0.999901 |

| Location | 18,209,403 – 18,209,510 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.40 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.88 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.46 |
| SVM RNA-class probability | 0.999901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18209403 107 + 22224390 UCAAAUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCGUC-------------CAUGGAACUGCUUCAUCGCCUAACUGCAGCAUAUGGACUCAAAUGAUUGUAAUU ...(((((.((((((.(((((.....((((....(((((((((((.(((..((-------------....)).))).)))))))))))....)))).....)))))..)))))).))))) ( -38.80) >DroSec_CAF1 69507 107 + 1 ACAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCGUC-------------CAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUGAACUCAAAUGCUUGUAAUU ........((((....(((((.....((((...((((((((((((.(((..((-------------....)).))).))))))))))))...)))).....)))))....))))...... ( -39.70) >DroSim_CAF1 68772 107 + 1 UCAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCGUC-------------CAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUGAACUCAAAUGCUUGUAAUC ........((((....(((((.....((((...((((((((((((.(((..((-------------....)).))).))))))))))))...)))).....)))))....))))...... ( -39.70) >DroEre_CAF1 63600 111 + 1 UCAACUUUAAGUUCUCAAGUUCUCGAGCUGGAUUUAGGCGAUGAAUCUGCGG-CACGUGGCGCAUUCAUGGGACUUAUUCAUCGUCUAUCUACAGCAUGCA-----GCAU--AUA-AAUU ..................(((..((.((((....((((((((((((.((((.-(....).)))).((....))...))))))))))))....)))).)).)-----))..--...-.... ( -28.50) >DroYak_CAF1 63349 108 + 1 UCAACUUGAAGUUCCCAUGCU-------GGGAUUUAGACGAUGAAUCAGCGGCCACUUGGAGCAUUAGUGGAACUGCUUCAUCGUCUAGCUUCAGCAUAUA---UUGAAU--UUACAAUU .....(((((((((..(((((-------(((..((((((((((((.(((...(((((.........)))))..))).)))))))))))).))))))))...---..))))--)).))).. ( -38.60) >consensus UCAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCGUC_____________CAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUG_ACUCAAAUG_UUGUAAUU ..(((((((.....))))))).....((((...((((((((((((.(((........................))).))))))))))))...))))........................ (-23.52 = -24.88 + 1.36)

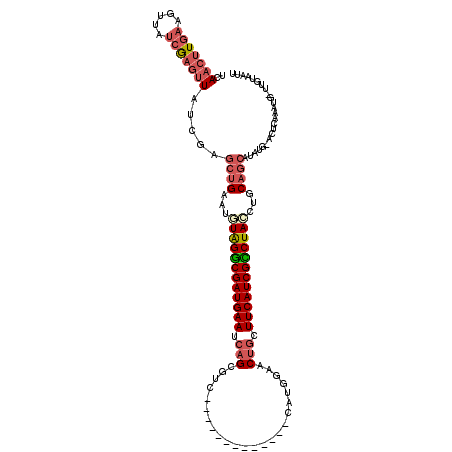

| Location | 18,209,403 – 18,209,510 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.40 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -19.91 |
| Energy contribution | -22.27 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.32 |
| SVM RNA-class probability | 0.999869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18209403 107 - 22224390 AAUUACAAUCAUUUGAGUCCAUAUGCUGCAGUUAGGCGAUGAAGCAGUUCCAUG-------------GACGCUGAUUCAUCGCCUACAUUCAGCUCGAUAACUCGAUAACUUCAAUUUGA ............((((((......((((.((((((((((((((.((((((....-------------)).)))).))))))))))).)))))))......)))))).............. ( -35.80) >DroSec_CAF1 69507 107 - 1 AAUUACAAGCAUUUGAGUUCAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUG-------------GACGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGU ......(((...(((((((.....((((...((.(((((((((.((((((....-------------)).)))).))))))))).))...)))).....)))))))...)))........ ( -36.60) >DroSim_CAF1 68772 107 - 1 GAUUACAAGCAUUUGAGUUCAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUG-------------GACGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGA ((((..(((...(((((((.....((((...((.(((((((((.((((((....-------------)).)))).))))))))).))...)))).....)))))))...)))..)))).. ( -36.80) >DroEre_CAF1 63600 111 - 1 AAUU-UAU--AUGC-----UGCAUGCUGUAGAUAGACGAUGAAUAAGUCCCAUGAAUGCGCCACGUG-CCGCAGAUUCAUCGCCUAAAUCCAGCUCGAGAACUUGAGAACUUAAAGUUGA ..((-(((--(.((-----.....))))))))(((.((((((((..((..((((.........))))-..))..)))))))).))).......(((((....)))))............. ( -23.40) >DroYak_CAF1 63349 108 - 1 AAUUGUAA--AUUCAA---UAUAUGCUGAAGCUAGACGAUGAAGCAGUUCCACUAAUGCUCCAAGUGGCCGCUGAUUCAUCGUCUAAAUCCC-------AGCAUGGGAACUUCAAGUUGA .((((...--...)))---)....(((((((.(((((((((((.((((.(((((.........)))))..)))).)))))))))))..((((-------.....)))).)))).)))... ( -37.00) >consensus AAUUACAA_CAUUUGAGU_CAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUG_____________GACGCUGAUUCAUCGCCUACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGA ............((((((......((((.(..(((((((((((.((((......................)))).)))))))))))..).))))......)))))).............. (-19.91 = -22.27 + 2.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:36 2006