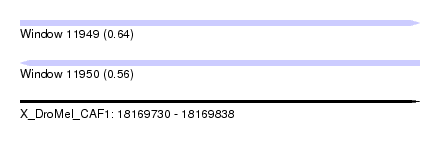
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,169,730 – 18,169,838 |
| Length | 108 |
| Max. P | 0.641663 |

| Location | 18,169,730 – 18,169,838 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -7.48 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
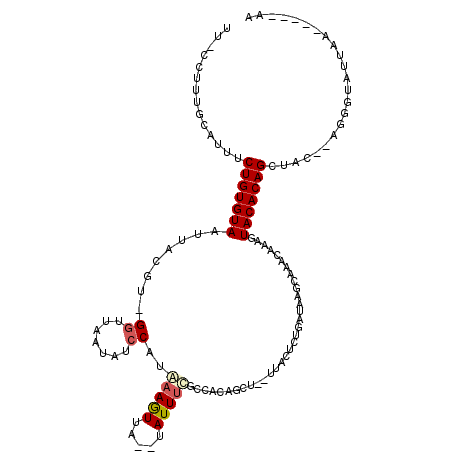
>X_DroMel_CAF1 18169730 108 + 22224390 UU----UUUAAUACCCU--GUAGCUGUGUACUUUAUUUACUUAUCAGACUAA--AGCUGUGGCCAAAUA--UCACUUUACGGAUAUUAACC-ACGUAAUUACACAGAAAUGCAAAGG-AA ..----.......((.(--(((.((((((((((((.((........)).)))--))..((((...((((--((........))))))..))-)).....)))))))...))))..))-.. ( -23.30) >DroSec_CAF1 31714 107 + 1 UU-----UUAAUACCCU--GUAGCUGUGUACUUUGUUUGCUUAUCAGAGUAA--AGCUGUGGCAAAAUA--UAACUUUAUGGAUAUUAACC-ACGUAAUUACACAGAAACGCAAAGG-AA ..-----.......(((--((..(((((((.(((((.....((((...((((--((.((((......))--)).)))))).))))......-))).)).)))))))..))....)))-.. ( -20.70) >DroSim_CAF1 31825 112 + 1 UU-----UUAAUACCCUUAGUAGCUGUGUACUUUGUUUGCUUAUCAGAGUAACUACCAUUCAAGGAAUA--UAACUUUAUGGAUAUUAACCAACGGAAUUACACAGAAACUCAGUGG-GA ..-----......(((..(((..(((((((.(((((((((((....)))))....((((..(((.....--...))).)))).........))))))..)))))))..)))....))-). ( -25.70) >DroEre_CAF1 27096 105 + 1 UUGUAUGUAAAUACCCU--GUAGCUGUGUAUU--------UUAACCGAGUAA--GGCUGCGGCGAAAUA--UCAUUUCAUGGAUAUUAACC-ACGUAAUUACACAGAAAUCCAAAUUGAA .....(((((.((((((--((((((...((((--------(.....))))).--)))))))).).((((--((........))))))....-..))).)))))................. ( -20.90) >DroYak_CAF1 29398 110 + 1 AU-----UAAAUACCCU--GUAGCUGUGUAUUCUGAAUAUUUAUCAGAGAAA--AUCUCUGGCAAAAUAUAUAACUUUUUGCAUAUUAACC-ACGCAAUUACACAGAAAUGCUGAGGAAA ..-----......(((.--(((.(((((((...((.........(((((...--..)))))((((((.........)))))).........-...))..)))))))...))).).))... ( -25.30) >consensus UU_____UUAAUACCCU__GUAGCUGUGUACUUUGUUUACUUAUCAGAGUAA__AGCUGUGGCAAAAUA__UAACUUUAUGGAUAUUAACC_ACGUAAUUACACAGAAAUGCAAAGG_AA ...................((..((((((((((((.........)))))...........................(((((............))))).)))))))..)).......... ( -7.48 = -8.40 + 0.92)



| Location | 18,169,730 – 18,169,838 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -6.44 |
| Energy contribution | -6.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18169730 108 - 22224390 UU-CCUUUGCAUUUCUGUGUAAUUACGU-GGUUAAUAUCCGUAAAGUGA--UAUUUGGCCACAGCU--UUAGUCUGAUAAGUAAAUAAAGUACACAGCUAC--AGGGUAUUAAA----AA ..-.(((((.....((((((......((-(((((((((((.....).))--)).)))))))).(((--(((..((....))....))))))))))))...)--)))).......----.. ( -26.30) >DroSec_CAF1 31714 107 - 1 UU-CCUUUGCGUUUCUGUGUAAUUACGU-GGUUAAUAUCCAUAAAGUUA--UAUUUUGCCACAGCU--UUACUCUGAUAAGCAAACAAAGUACACAGCUAC--AGGGUAUUAA-----AA .(-(((....((..(((((((.....((-(((.(((((..........)--))))..))))).(((--(.........))))........)))))))..))--))))......-----.. ( -22.10) >DroSim_CAF1 31825 112 - 1 UC-CCACUGAGUUUCUGUGUAAUUCCGUUGGUUAAUAUCCAUAAAGUUA--UAUUCCUUGAAUGGUAGUUACUCUGAUAAGCAAACAAAGUACACAGCUACUAAGGGUAUUAA-----AA .(-((....(((..(((((((......(((.(((....((((.(((...--.....)))..))))(((.....))).))).)))......)))))))..)))..)))......-----.. ( -22.80) >DroEre_CAF1 27096 105 - 1 UUCAAUUUGGAUUUCUGUGUAAUUACGU-GGUUAAUAUCCAUGAAAUGA--UAUUUCGCCGCAGCC--UUACUCGGUUAA--------AAUACACAGCUAC--AGGGUAUUUACAUACAA .....((((.....(((((((.....((-(((.((((((........))--))))..)))))((((--......))))..--------..)))))))...)--)))((((....)))).. ( -23.80) >DroYak_CAF1 29398 110 - 1 UUUCCUCAGCAUUUCUGUGUAAUUGCGU-GGUUAAUAUGCAAAAAGUUAUAUAUUUUGCCAGAGAU--UUUCUCUGAUAAAUAUUCAGAAUACACAGCUAC--AGGGUAUUUA-----AU ..((((.(((.....((((((.((((((-(.....)))))))........((((((...(((((..--...)))))..))))))......)))))))))..--))))......-----.. ( -24.50) >consensus UU_CCUUUGCAUUUCUGUGUAAUUACGU_GGUUAAUAUCCAUAAAGUUA__UAUUUCGCCACAGCU__UUACUCUGAUAAGCAAACAAAGUACACAGCUAC__AGGGUAUUAA_____AA ..............(((((((........((.......))..(((((.....))))).................................)))))))....................... ( -6.44 = -6.92 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:21 2006