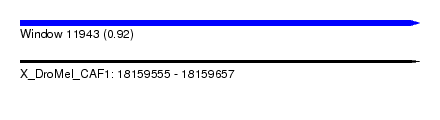
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,159,555 – 18,159,657 |
| Length | 102 |
| Max. P | 0.916135 |

| Location | 18,159,555 – 18,159,657 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.87 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.33 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18159555 102 + 22224390 -------UGUAGACA----UGCUACAUGGCUCCAUUGCCAUAU-UGGCAAUGCAUCAACAAGUUCUCCUUUACUUCUUUAUCCCAGUGA--CUUACUCCCGCAGUG----GGACCUGCUU -------.((((...----(((((.(((((......)))))..-)))))...............................(((((.((.--(........))).))----))).)))).. ( -24.90) >DroSec_CAF1 21662 105 + 1 -------UGUAGACAUGUAUGCUACAUGGUUCCAUUGCCAUAU-UGGCAAUGCAUCAACAAGUUCUCCUUUACUUCUUUAUCCCGG-GA--AUUACUCCCUCAGUG----GGACCCGCUU -------(((((.((....))))))).((((((((((((....-.))))))((......((((........)))).........((-((--.....))))...)))----)))))..... ( -27.10) >DroSim_CAF1 21706 105 + 1 -------UGUAGACAUGUAUGCUACAUGGUUCCAUUGCCAUAU-UGGCAAUGCAUCAACAAGUUCUCCUUUACUUCUUUAUCCCGG-GA--CUUACUCCCUCAGUG----GGACCCGCUU -------(((((.((....)))))))((((..(((((((....-.))))))).))))..((((........))))........(((-(.--((((((.....))))----)).))))... ( -27.80) >DroEre_CAF1 17884 100 + 1 -------GAUAUACAUGUA-----GGUGGUUCCAUUGCCAUAU-UGGCAAUGCAUCAACAAGUCCCCCUUUACAUCUUUAUUAAGG-GA--CUUACUCCUUCAACG----GGACCCGCUU -------...........(-----(((((((((((((((....-.))))))........(((((((..................))-))--)))...........)----))).)))))) ( -27.57) >DroYak_CAF1 19875 104 + 1 -------GAUAUACAUGUA-----GGUGGUUCCAUUGCCGCAU-UGGCAAUGCAUCAACAAGUACUGCUUUUCGUCAUUUUUCCGG-GC--UUUACUCCCUUACUAGGAAGGACCCGCUU -------....(((.(((.-----((((....(((((((....-.))))))))))).))).)))...................(((-((--(((....(((....))))))).))))... ( -25.10) >DroAna_CAF1 65407 102 + 1 UAUAUUUGGUAUAUAAGUA---------UGUCUGUUGCAAUUUAUAGCAACUUAUCAAAAAAUCCCCAUCCUGGCCUCUAUUGGUG-GAUCCUUAAUCCGGAUAUC----GGC----CCU ....(((((((....((..---------...))(((((........))))).))))))).............((((..((((..((-(((.....)))))))))..----)))----).. ( -20.30) >consensus _______GGUAGACAUGUA_____CAUGGUUCCAUUGCCAUAU_UGGCAAUGCAUCAACAAGUUCUCCUUUACUUCUUUAUCCCGG_GA__CUUACUCCCUCAGUG____GGACCCGCUU ...........................(((((((((((((....))))))))................................((.((.......))))..........)))))..... (-11.23 = -11.33 + 0.10)

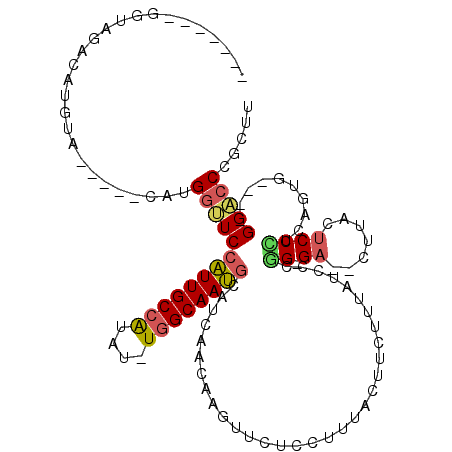

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:15 2006