
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,154,107 – 18,154,211 |
| Length | 104 |
| Max. P | 0.729366 |

| Location | 18,154,107 – 18,154,211 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.81 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
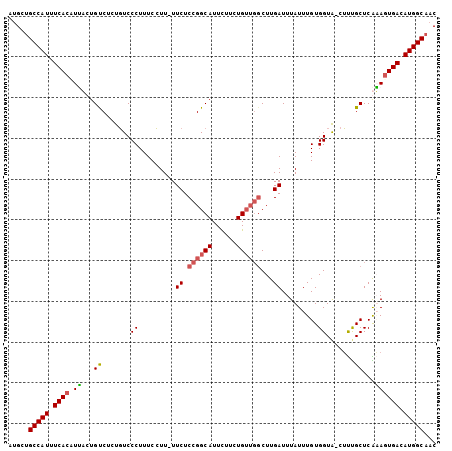
>X_DroMel_CAF1 18154107 104 + 22224390 AUGCUGCCAUUUCACAUAACUGCCUCUGUCCCUUUCCUU-UUCUCCGGCAUUCUUUUGUUGGCUUGAUUUAUUUGUGGUAUCUUCGCUCGUAUUGACAUGGCAAC ....((((((.(((.(((...((....((.((.......-.((.((((((......))))))...)).........)).))....))...)))))).)))))).. ( -20.83) >DroSec_CAF1 16337 103 + 1 AUGCUGCCAUUUCACAUUUCUGUCUCUGUCCCUUUCCUU-UUCUCCGGCAUUUUUCUGUUGGCUUGAUUUAUUUGUGGUA-CUUUGCUCAAAGUGACAUGGCAAC ....((((((.((((.(((..((....((.((.......-.((.((((((......))))))...)).........)).)-)...))..))))))).)))))).. ( -22.73) >DroSim_CAF1 16366 103 + 1 AUGCUGCCAUUUCACAUUUCUGUCUCUGUCCCUUUCCUU-UUCUCCGGCAUUUCUCUGUUGGCUUGAUUUAUUUGUGGUA-CUUUGCUCAAAGUGACAUGGCAAC ....((((((.((((.(((..((....((.((.......-.((.((((((......))))))...)).........)).)-)...))..))))))).)))))).. ( -22.73) >DroEre_CAF1 12651 100 + 1 AAGCAGCCAUUUCACUUGACUGUCUCUCUCCCUUUCGUUCUUCUCCGGCAUUCUUCUG----CUUGAUUUAUUUGUGGUG-CGCUGCUCGUAGUGACAUGGCAAC .....(((((.((((((((........................((.((((......))----)).)).......(..(..-..)..)))).))))).)))))... ( -21.90) >consensus AUGCUGCCAUUUCACAUUACUGUCUCUGUCCCUUUCCUU_UUCUCCGGCAUUCUUCUGUUGGCUUGAUUUAUUUGUGGUA_CUUUGCUCAAAGUGACAUGGCAAC .....(((((.((((.((...((.......((.........((.((((((......))))))...)).........)).......))...)))))).)))))... (-16.31 = -16.81 + 0.50)


| Location | 18,154,107 – 18,154,211 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
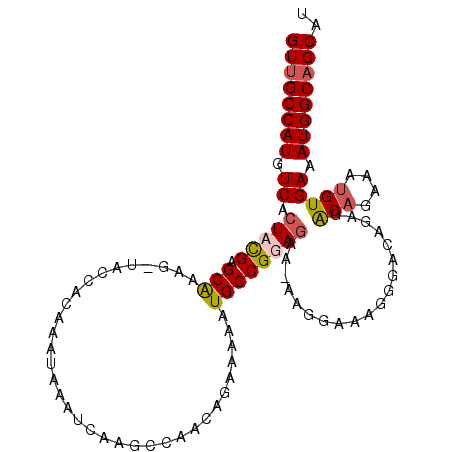
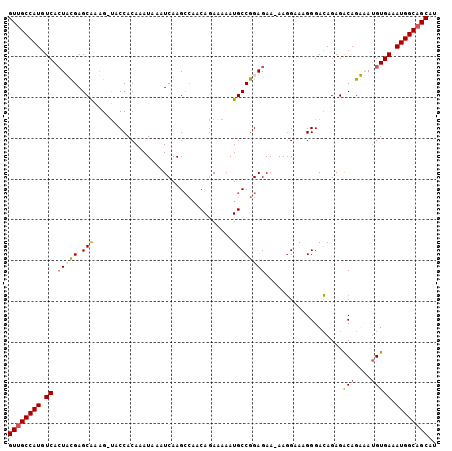
>X_DroMel_CAF1 18154107 104 - 22224390 GUUGCCAUGUCAAUACGAGCGAAGAUACCACAAAUAAAUCAAGCCAACAAAAGAAUGCCGGAGAA-AAGGAAAGGGACAGAGGCAGUUAUGUGAAAUGGCAGCAU ((((((((.(((.....(((...(((...........)))..(((............((......-..))...........))).)))...))).)))))))).. ( -22.50) >DroSec_CAF1 16337 103 - 1 GUUGCCAUGUCACUUUGAGCAAAG-UACCACAAAUAAAUCAAGCCAACAGAAAAAUGCCGGAGAA-AAGGAAAGGGACAGAGACAGAAAUGUGAAAUGGCAGCAU ((((((((((((((((....))))-).((..................((......))((......-..))...)))))....(((....)))...)))))))).. ( -23.00) >DroSim_CAF1 16366 103 - 1 GUUGCCAUGUCACUUUGAGCAAAG-UACCACAAAUAAAUCAAGCCAACAGAGAAAUGCCGGAGAA-AAGGAAAGGGACAGAGACAGAAAUGUGAAAUGGCAGCAU ((((((((((((((((....))))-).((..................((......))((......-..))...)))))....(((....)))...)))))))).. ( -23.00) >DroEre_CAF1 12651 100 - 1 GUUGCCAUGUCACUACGAGCAGCG-CACCACAAAUAAAUCAAG----CAGAAGAAUGCCGGAGAAGAACGAAAGGGAGAGAGACAGUCAAGUGAAAUGGCUGCUU ((.(((((.(((((((......(.-(.((.........((..(----((......)))..))......(....))).).).....))..))))).))))).)).. ( -22.60) >consensus GUUGCCAUGUCACUACGAGCAAAG_UACCACAAAUAAAUCAAGCCAACAGAAAAAUGCCGGAGAA_AAGGAAAGGGACAGAGACAGAAAUGUGAAAUGGCAGCAU ((((((((.((.(((((.(((..................................))))))))...................(((....))))).)))))))).. (-16.68 = -17.30 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:14 2006