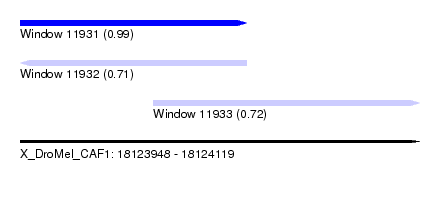
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,123,948 – 18,124,119 |
| Length | 171 |
| Max. P | 0.987845 |

| Location | 18,123,948 – 18,124,045 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.04 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18123948 97 + 22224390 GGAUCGACGGCGAUCGGUGAUCGAAGAUCGAAGAUCGAAGAUCGGAGAUGGCAGAUCGGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCA (((((..(..(((((...)))))..).(((((..(((((((((...(((.....)))...)))))))))..)))))))))).(((((.....))))) ( -36.70) >DroSec_CAF1 6503 76 + 1 GGAUCGGCGGCGAUCGGUGAUCGAAGAUCG---------------------CAGAUCGGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCA (((((.....(((((.((((((...)))))---------------------).)))))(((.((....)).)))..))))).(((((.....))))) ( -29.50) >DroSim_CAF1 7160 76 + 1 GGAUCGGCGGCGAUCGGUGAUCGAAGAUCG---------------------CAGAUCGGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCA (((((.....(((((.((((((...)))))---------------------).)))))(((.((....)).)))..))))).(((((.....))))) ( -29.50) >DroEre_CAF1 6326 76 + 1 GGAUCGGCGGCGAUCGGUGAUCGCAGAUCG---------------------GAGAUCGGAGAUCUUCGGAGCUCGAGAUCCGUGCUACACAUUAGCA ((((((((.((((((...))))))...(((---------------------((((((...))))))))).)))...))))).(((((.....))))) ( -30.50) >consensus GGAUCGGCGGCGAUCGGUGAUCGAAGAUCG_____________________CAGAUCGGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCA (((((..((((..((((.((((...((((........................))))...)))).)))).)).)).))))).(((((.....))))) (-26.22 = -26.04 + -0.19)
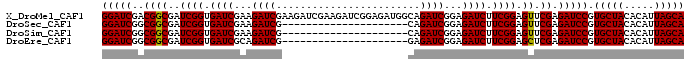


| Location | 18,123,948 – 18,124,045 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18123948 97 - 22224390 UGCUAAUGUGUAGCACGGAUCUCGAACUCCGAAGAUCUCCGAUCUGCCAUCUCCGAUCUUCGAUCUUCGAUCUUCGAUCACCGAUCGCCGUCGAUCC (((((.....))))).((...(((((...((((((((...((((..........))))...))))))))...)))))...))(((((....))))). ( -28.40) >DroSec_CAF1 6503 76 - 1 UGCUAAUGUGUAGCACGGAUCUCGAACUCCGAAGAUCUCCGAUCUG---------------------CGAUCUUCGAUCACCGAUCGCCGCCGAUCC (((((.....))))).((((((((.....)).))))))..((((.(---------------------((.....(((((...))))).))).)))). ( -21.50) >DroSim_CAF1 7160 76 - 1 UGCUAAUGUGUAGCACGGAUCUCGAACUCCGAAGAUCUCCGAUCUG---------------------CGAUCUUCGAUCACCGAUCGCCGCCGAUCC (((((.....))))).((((((((.....)).))))))..((((.(---------------------((.....(((((...))))).))).)))). ( -21.50) >DroEre_CAF1 6326 76 - 1 UGCUAAUGUGUAGCACGGAUCUCGAGCUCCGAAGAUCUCCGAUCUC---------------------CGAUCUGCGAUCACCGAUCGCCGCCGAUCC (((((.....))))).(((..(((.((..((.(((((...))))).---------------------))....((((((...)))))).)))))))) ( -22.20) >consensus UGCUAAUGUGUAGCACGGAUCUCGAACUCCGAAGAUCUCCGAUCUG_____________________CGAUCUUCGAUCACCGAUCGCCGCCGAUCC (((((.....))))).(((((.((......(((((((...............................)))))))((((...))))..))..))))) (-18.33 = -18.58 + 0.25)


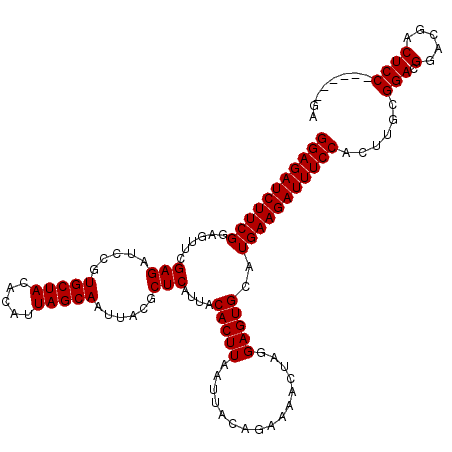
| Location | 18,124,005 – 18,124,119 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.62 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -27.97 |
| Energy contribution | -27.97 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18124005 114 + 22224390 GGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCC------GA (((((((((((......(((.....(((((.....)))))......)))....(((((................)))))..))))))))))).....((((.(.....))))------). ( -30.69) >DroSec_CAF1 6539 114 + 1 GGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCC------GA (((((((((((......(((.....(((((.....)))))......)))....(((((................)))))..))))))))))).....((((.(.....))))------). ( -30.69) >DroSim_CAF1 7196 114 + 1 GGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCC------GA (((((((((((......(((.....(((((.....)))))......)))....(((((................)))))..))))))))))).....((((.(.....))))------). ( -30.69) >DroEre_CAF1 6362 120 + 1 GGAGAUCUUCGGAGCUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCCGACUCCGA (((((((((((..(((((((.....(((((.....)))))......)))...............((....))..))))...))))))))))).....((((((((....))))..)))). ( -37.50) >DroYak_CAF1 6904 120 + 1 GGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCCGACUCCGA (((((((((((......(((.....(((((.....)))))......)))....(((((................)))))..))))))))))).....((((((((....))))..)))). ( -34.99) >consensus GGAGAUCUUCGGAGUUCGAGAUCCGUGCUACACAUUAGCAAUUACGCUCAUUACACUUAAUUACAGAAAACUAGGAGUGCAUGAAGAUUUCCACUUGCGGACGGACGACUCC______GA (((((((((((......(((.....(((((.....)))))......)))....(((((................)))))..)))))))))))......(((.(.....))))........ (-27.97 = -27.97 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:06 2006