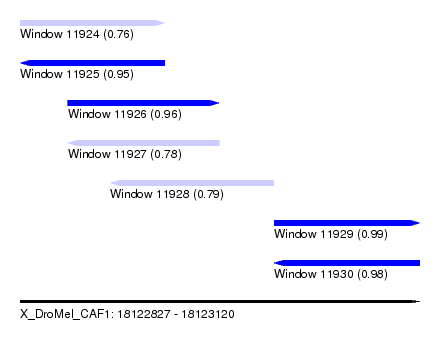
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,122,827 – 18,123,120 |
| Length | 293 |
| Max. P | 0.986640 |

| Location | 18,122,827 – 18,122,933 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -29.27 |
| Energy contribution | -29.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18122827 106 + 22224390 GGGAGUUGGAUGGCAUGGCGAAGGGGCGGGGGAAAAGGGAAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCU ............((....(....).))..............(((((...(((((....(((((.......)))))...)))))(((((.....)))))...))))) ( -29.10) >DroSec_CAF1 5435 99 + 1 GGGAGGUGGACGGGAUGGUGAAGGGCCAGGGG-------AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCU (((.(((...((.((((.(((.((((((..((-------((..(.((...(((....)))....)).)..))).)...)))))).)))..))))..))..)))))) ( -32.10) >DroSim_CAF1 6097 99 + 1 GGGAGGUGGACGGGAUGGUGAAGGGCCAGGGG-------AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCU (((.(((...((.((((.(((.((((((..((-------((..(.((...(((....)))....)).)..))).)...)))))).)))..))))..))..)))))) ( -32.10) >consensus GGGAGGUGGACGGGAUGGUGAAGGGCCAGGGG_______AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCU ...............((((.....)))).............(((((...(((((....(((((.......)))))...)))))(((((.....)))))...))))) (-29.27 = -29.17 + -0.11)

| Location | 18,122,827 – 18,122,933 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.25 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18122827 106 - 22224390 AGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUUCCCUUUUCCCCCGCCCCUUCGCCAUGCCAUCCAACUCCC ((((((..(((((.....)))))((((....((((.........)))).....))))......................))))))..................... ( -23.90) >DroSec_CAF1 5435 99 - 1 AGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU-------CCCCUGGCCCUUCACCAUCCCGUCCACCUCCC .((((..((((((.....))))))..)....(((((..(((....))).....((((..........-------.....)))).)))))...)))........... ( -27.36) >DroSim_CAF1 6097 99 - 1 AGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU-------CCCCUGGCCCUUCACCAUCCCGUCCACCUCCC .((((..((((((.....))))))..)....(((((..(((....))).....((((..........-------.....)))).)))))...)))........... ( -27.36) >consensus AGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU_______CCCCUGGCCCUUCACCAUCCCGUCCACCUCCC .(((((.((((((.....)))))))))....(((((..(((....))).....((((......................)))).)))))....))........... (-23.14 = -23.25 + 0.11)



| Location | 18,122,862 – 18,122,973 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -33.33 |
| Energy contribution | -33.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18122862 111 + 22224390 AGGGAAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCUUUCGGACAAAAGGGCCAGGACAAAACUUCACAGGCGAAUG ...((((((((...(((((....(((((.......)))))...)))))(((((.....)))))...))))))))..........(((..((.......))...)))..... ( -37.70) >DroSec_CAF1 5467 107 + 1 ----AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCUUUCGGACAAAAGGGCCAGGACAAAACUUCACAGGCGAAUG ----(((((((...(((((....(((((.......)))))...)))))(((((.....)))))...)))))))...........(((..((.......))...)))..... ( -33.20) >DroSim_CAF1 6129 107 + 1 ----AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCUUUCGGACAAAAGGGCCAGGACAAAACUUCACAGGCGAAUG ----(((((((...(((((....(((((.......)))))...)))))(((((.....)))))...)))))))...........(((..((.......))...)))..... ( -33.20) >consensus ____AAAGGGGAAAGGCCGAAGGGUGAGGUCACAAUUCACAAGUGGCCCGUCAAAUAUUGACGGAACCCCUUUCGGACAAAAGGGCCAGGACAAAACUUCACAGGCGAAUG ....(((((((...(((((....(((((.......)))))...)))))(((((.....)))))...)))))))...........(((..((.......))...)))..... (-33.33 = -33.33 + 0.00)


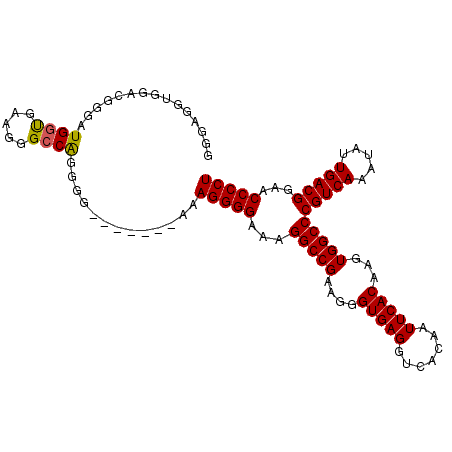
| Location | 18,122,862 – 18,122,973 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18122862 111 - 22224390 CAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUUCCCU .....(((.(.((.......))).)))..........((((((((...(((((.....)))))((((....((((.........)))).....))))...))))))))... ( -31.60) >DroSec_CAF1 5467 107 - 1 CAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU---- .....((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))......((((..(((....)))..)))).............---- ( -30.50) >DroSim_CAF1 6129 107 - 1 CAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU---- .....((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))......((((..(((....)))..)))).............---- ( -30.50) >consensus CAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGUGACCUCACCCUUCGGCCUUUCCCCUUU____ .....((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))......((((..(((....)))..))))................. (-30.50 = -30.50 + 0.00)



| Location | 18,122,893 – 18,123,013 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -37.14 |
| Energy contribution | -37.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18122893 120 - 22224390 GGAAGGGAGCGCGAUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU ((..((((......))))....((((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... ( -37.50) >DroSec_CAF1 5494 120 - 1 GGAAGGGAGCGCGAUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU ((..((((......))))....((((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... ( -37.50) >DroSim_CAF1 6156 120 - 1 GGAAGGGAGCGCGAUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU ((..((((......))))....((((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... ( -37.50) >DroEre_CAF1 5242 120 - 1 GGUAGGGAGCGGGAUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU (((.((((......)))).....(((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... ( -37.30) >DroYak_CAF1 5634 120 - 1 GGAAGGGAGCGGGUUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU ((..(((((....)))))....((((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... ( -38.30) >consensus GGAAGGGAGCGCGAUUCCAUAUUCAAAUAUUUGACCAUCGCAUUCGCCUGUGAAGUUUUGUCCUGGCCCUUUUGUCCGAAAGGGGUUCCGUCAAUAUUUGACGGGCCACUUGUGAAUUGU ((..((((......))))....((((....)))))).(((((...((((((.((((.(((.(..(((((((((.....)))))))))..).))).)))).))))))....)))))..... (-37.14 = -37.14 + 0.00)



| Location | 18,123,013 – 18,123,120 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.12 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18123013 107 + 22224390 UC-CCCCGCUCCCUUUUAAGGGAUGUGGAC------------GUGGAUGUGGAAUGCGGUGUCGAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGUAGUCGCAUUCG ((-(((((((((((....))))).))))..------------).)))....(((((((((((((...........)))))))....((((((..(((....)))..))))).))))))). ( -36.50) >DroSec_CAF1 5614 108 + 1 UUCCGCCGCUCCCUUCUAAGGGAUGUGGAC------------GUGGAUGUGGAAUGCGGUGUCAAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGUAGUCGCAUUCG ((((((((((((((....))))).)))).)------------).)))....(((((((((((((...........)))))))....((((((..(((....)))..))))).))))))). ( -39.10) >DroSim_CAF1 6276 107 + 1 UC-CCCCGCUCCCUUUUAAGGGAUGUGGAC------------GUAGAUGUGGAAUGCGGUGUCAAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGUAGUCGCAUUCG ..-..(((((((((....))))).))))..------------.........(((((((((((((...........)))))))....((((((..(((....)))..))))).))))))). ( -36.10) >DroEre_CAF1 5362 119 + 1 UC-CCCCGCUCCCUUUUAAGGGAUGUGGAUGUGGAUGUGGAUGUGGAUGUGGAAUGCGGUGUCGAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGGAGUCGCAUUCG ((-(((((((((((....))))).))))..).)))...((((((((...(.((.((((((((((...........)))))))....(((((....))))).))).)).)..)))))))). ( -39.30) >DroYak_CAF1 5754 113 + 1 UC-CCCCGCUCCCUUUUAAGGGAUGUGGAUGUG------GAUGUGGAUGUGGAAUGCGGUGUCGAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGGAGUCGCAUUCG ..-..(((((((((....))))).))))....(------(((((((...(.((.((((((((((...........)))))))....(((((....))))).))).)).)..)))))))). ( -38.20) >consensus UC_CCCCGCUCCCUUUUAAGGGAUGUGGAC____________GUGGAUGUGGAAUGCGGUGUCGAUCUUGUUAACUGAUAUCAUCCGUUGCAGUUGCAACUGCAGUUGUAGUCGCAUUCG .....(((((((((....))))).))))................((((((((..((((((((((...........)))))))....(((((....))))).))).......)))))))). (-35.16 = -35.12 + -0.04)



| Location | 18,123,013 – 18,123,120 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18123013 107 - 22224390 CGAAUGCGACUACAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUCGACACCGCAUUCCACAUCCAC------------GUCCACAUCCCUUAAAAGGGAGCGGGG-GA .(((((((.........(((((....)))))(((.(((...))).)))..............)))))))........(------------.(((.(.(((((....)))))).))).-). ( -28.80) >DroSec_CAF1 5614 108 - 1 CGAAUGCGACUACAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUUGACACCGCAUUCCACAUCCAC------------GUCCACAUCCCUUAGAAGGGAGCGGCGGAA .(((((((........((((((....))))))..........((((((.....))))))...)))))))........(------------(.((.(.(((((....)))))).))))... ( -31.80) >DroSim_CAF1 6276 107 - 1 CGAAUGCGACUACAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUUGACACCGCAUUCCACAUCUAC------------GUCCACAUCCCUUAAAAGGGAGCGGGG-GA .(((((((........((((((....))))))..........((((((.....))))))...)))))))........(------------.(((.(.(((((....)))))).))).-). ( -32.10) >DroEre_CAF1 5362 119 - 1 CGAAUGCGACUCCAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUCGACACCGCAUUCCACAUCCACAUCCACAUCCACAUCCACAUCCCUUAAAAGGGAGCGGGG-GA .(((((((..(((...((((((....))))))..)))((((....(....)...))))....)))))))................(((...(((.(.(((((....)))))).))))-)) ( -30.30) >DroYak_CAF1 5754 113 - 1 CGAAUGCGACUCCAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUCGACACCGCAUUCCACAUCCACAUC------CACAUCCACAUCCCUUAAAAGGGAGCGGGG-GA .(((((((..(((...((((((....))))))..)))((((....(....)...))))....)))))))..........((------(...(((.(.(((((....)))))).))))-)) ( -30.30) >consensus CGAAUGCGACUACAACUGCAGUUGCAACUGCAACGGAUGAUAUCAGUUAACAAGAUCGACACCGCAUUCCACAUCCAC____________GUCCACAUCCCUUAAAAGGGAGCGGGG_GA .((((((.........((((((....))))))..((.((..(((.........)))...))))))))))......................(((.(.(((((....)))))).))).... (-26.22 = -26.42 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:04 2006