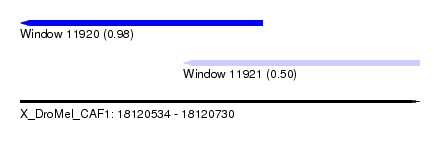
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,120,534 – 18,120,730 |
| Length | 196 |
| Max. P | 0.980564 |

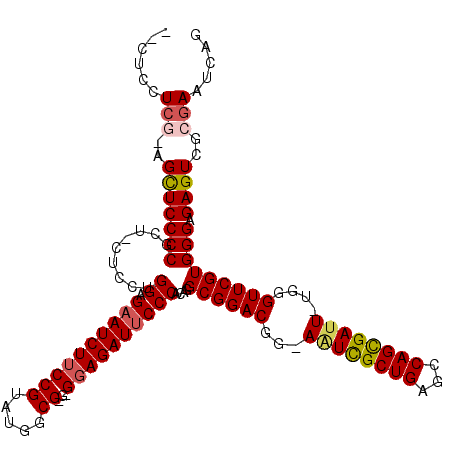
| Location | 18,120,534 – 18,120,653 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -39.84 |
| Energy contribution | -40.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18120534 119 - 22224390 GAGCCAGCGAUU-UAGGUUCGUGGGAGAGUCGCGAAUCAGAAACUCCACGCGAAUCGUUUUACAAUCGGAACUAUUGGAGUUACUGGACACUGGACACACGCCCUGGUGGGCGGUCUUCG ..(((((..(((-(.((((((((((((..((........))..)))).))))))))(((((......))))).....))))..)))).)...(((.((.(((((....))))))).))). ( -43.60) >DroSec_CAF1 3152 119 - 1 GAGCCAGCGAUU-UGGGUUCGUGGGAGAGUCGGGAAUCAGAAACUCCACGCGAAUCGUUUUACAAUCGGAACUAUGGGAGUUACUGGACACUGGACACACGCCCUGGUGGGCGGUCUUUG ..(((((..(((-(.((((((((((((..((........))..)))).))))))))(((((......))))).....))))..)))).)...((((...(((((....)))))))))... ( -42.20) >DroSim_CAF1 3426 119 - 1 GAGCCAGCGAUU-UGGGUUCGUGGGAGAGUCGCGAAUCAGAAACUCCACGCGAAUCGUUUUACAAUCGGAACUAUUGGAGUUACUGGACACUGGACACACGCCCUGGUGGGCGGUCUUCG ..(((((..(((-(.((((((((((((..((........))..)))).))))))))(((((......))))).....))))..)))).)...(((.((.(((((....))))))).))). ( -43.40) >DroEre_CAF1 3044 119 - 1 GAGCCAGUGAUU-UCGGUUCGUGGGAGAGUCGCGAAUGAGAAUCUCCACGCGAAUCGUUUUACAAUCGGAACUAUUGGAGUUACUGGACACUGGACACACGCCCUGGUGGGCGGUCUCAC ..((((((((((-(((((((((((((((.((........)).))))).))))))))(((((......)))))....))))))))))).)....((.((.(((((....))))))).)).. ( -51.10) >DroYak_CAF1 3407 120 - 1 GAGCCAGCGAUUUUUGGUUCGUGGGAGAGUCGCGAAUCAGAAACUCCACGCGAAUCGUUUUACAAUCGGAACUAUUGCAGAUACUGGACACUGGCCACACGCCCUGGUGGGCGGUCUCCC (.((((((((((..(((((((((((((..((........))..)))).)))))))))......))))).........(((...)))....))))))...(((((....)))))....... ( -41.60) >consensus GAGCCAGCGAUU_UGGGUUCGUGGGAGAGUCGCGAAUCAGAAACUCCACGCGAAUCGUUUUACAAUCGGAACUAUUGGAGUUACUGGACACUGGACACACGCCCUGGUGGGCGGUCUUCG ..(((((.(......((((((((((((..((........))..)))).)))))))).........((((((((.....)))).)))).).)))).)...(((((....)))))....... (-39.84 = -40.04 + 0.20)

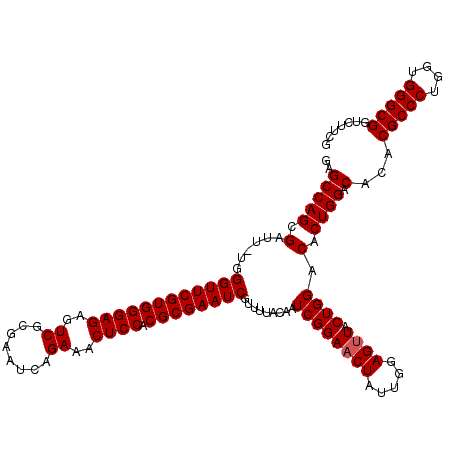
| Location | 18,120,614 – 18,120,730 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -48.66 |
| Consensus MFE | -34.92 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18120614 116 - 22224390 AGCUCCUCG-AGUUCCCCGCA-CUCCUGGAGAAUCUUCCGUAUGGCGCG-GGAGAUUCCCACAGGCGGACGGCAAUCGCUGAGCCAGCGAUU-UAGGUUCGUGGGAGAGUCGCGAAUCAG .....(.((-(.(..(((((.-..(((((.(((((((((((.....)))-)))))))).).)))).((((...((((((((...))))))))-...)))))))))..).))).)...... ( -50.30) >DroSec_CAF1 3232 114 - 1 --CUCCUCGGCGCUCCCCGCU-CUCCUGGUGAAUCUUCCGUAUGGCG-G-GGAGAUUCCCACAGGCGGACGGAAAUCGCUGAGCCAGCGAUU-UGGGUUCGUGGGAGAGUCGGGAAUCAG --...........((((.(((-(((((((.(((((((((........-)-))))))))))....((((((..(((((((((...))))))))-)..)))))))))))))).))))..... ( -55.50) >DroSim_CAF1 3506 114 - 1 --CUCCUCGGCGCUCCCCGCU-CUCCUGGUGAAUCUUCCGUAUGGCG-G-GGAGAUUCCCACAGGCGGACGGAAAUCGCUGAGCCAGCGAUU-UGGGUUCGUGGGAGAGUCGCGAAUCAG --.......(((((((((((.-..(((((.(((((((((........-)-)))))))).).)))).((((..(((((((((...))))))))-)..))))))))).))).)))....... ( -52.00) >DroEre_CAF1 3124 112 - 1 --CUCCUCG-AGCUCCCCUUU-CUCCCGGAGAAUCUGCCGUUUGGCG-A-GGAGAUUCCCACAGGCGGACGG-AUUUGCUGAGCCAGUGAUU-UCGGUUCGUGGGAGAGUCGCGAAUGAG --...((((-.((......((-(((((((.((((((.(((.....))-.-).))))))))....((((((.(-(.(..(((...)))..)..-)).)))))))))))))..))...)))) ( -41.40) >DroYak_CAF1 3487 115 - 1 --CUUUUGA-AGCUCCCCUUCGCUCCUGGAGCAUCUUCCGUAUGGCG-GUGGAGAUUCCCACAGGCGGACGG-AGUCGCUGAGCCAGCGAUUUUUGGUUCGUGGGAGAGUCGCGAAUCAG --..((((.-.((((..((((((((((((((....)))))...)).)-))))))..((((((..((.((..(-((((((((...))))))))))).))..))))))))))..)))).... ( -44.10) >consensus __CUCCUCG_AGCUCCCCGCU_CUCCUGGAGAAUCUUCCGUAUGGCG_G_GGAGAUUCCCACAGGCGGACGG_AAUCGCUGAGCCAGCGAUU_UGGGUUCGUGGGAGAGUCGCGAAUCAG ......(((..(((((((.........((.((((((((((.....))...))))))))))....((((((...((((((((...))))))))....))))))))).))))..)))..... (-34.92 = -35.52 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:56 2006