
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,119,551 – 18,119,659 |
| Length | 108 |
| Max. P | 0.766851 |

| Location | 18,119,551 – 18,119,659 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
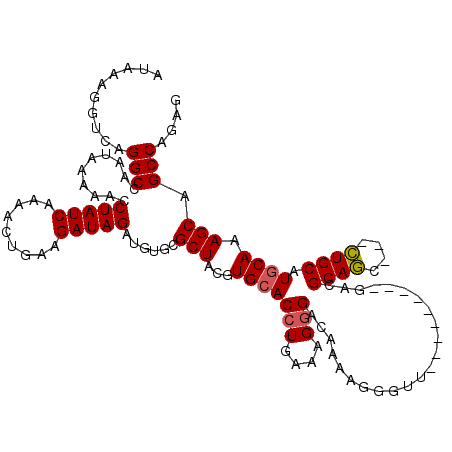
>X_DroMel_CAF1 18119551 108 + 22224390 CUCUGGCUGCUUUGCAUGCAG---GCUGCGUC---------AACCCUUUUGUCCCUUUCAGGUGCACGUAGCGCACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.(((....)))---(((((((.---------.(((...............)))..)))))))))..(((((((.........))))))).......)))).))....... ( -26.16) >DroSec_CAF1 2165 108 + 1 CUCUGGCUGCUUUGCAUGCAG---GCUGCGUC---------AACCCUUUUGUCUCUUUCAGGUCCACGUAGCGAACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.......))..---(((((((.---------.(((...............)))..)))))))(((.(((((((.........))))))).)))...)))).))....... ( -26.46) >DroSim_CAF1 2442 108 + 1 CUCUGGCUGCUUUGCAUGCAG---GCUGCGUC---------AACCCUUUUGUCCCUUUCAGGUGCACGUAGCGCACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.(((....)))---(((((((.---------.(((...............)))..)))))))))..(((((((.........))))))).......)))).))....... ( -26.16) >DroEre_CAF1 2095 117 + 1 CUCUGGCCGCUUUGCAUGCAG---GCUGCGUCAACUGCGUCAACCCUUUUGUCCCUUUCAGGUGCACGUAGCGCACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.(((....)))---(((((((((.(((.(.(((.....))).).....))).)).)))))))))..(((((((.........))))))).......)))).))....... ( -30.20) >DroYak_CAF1 2369 113 + 1 CUCUGGCUGCUUUGCAUGCAUCACAAUGCGUCA-------CAACCCUUUUGUCCCUUUCAGGUGCACGUAGCGCACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.((((((((((....))))))..-------.........((..((.....))..)).)))).))..(((((((.........))))))).......)))).))....... ( -25.00) >consensus CUCUGGCUGCUUUGCAUGCAG___GCUGCGUC_________AACCCUUUUGUCCCUUUCAGGUGCACGUAGCGCACAUCUAUCUUCAGUUUUGAUAGGUUUUUAUUGGCCUGACCUUUAU .((.((((((.((((((((((....))))))..................((..((.....))..)).)))).))..(((((((.........))))))).......)))).))....... (-24.66 = -24.22 + -0.44)


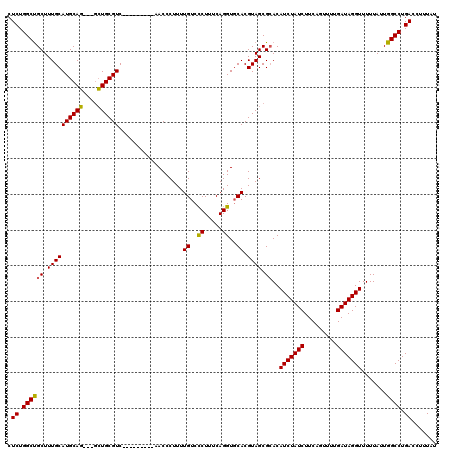
| Location | 18,119,551 – 18,119,659 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18119551 108 - 22224390 AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUGCGCUACGUGCACCUGAAAGGGACAAAAGGGUU---------GACGCAGC---CUGCAUGCAAAGCAGCCAGAG ..........(((..........(((((.........)))))...(((....(((((.(((....)))......(((((---------(...))))---)))))))....)))))).... ( -28.20) >DroSec_CAF1 2165 108 - 1 AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUUCGCUACGUGGACCUGAAAGAGACAAAAGGGUU---------GACGCAGC---CUGCAUGCAAAGCAGCCAGAG .....(((...((..........(((((.........)))))((((..(((.(((.(((((.............)))))---------.))).)))---..)))).....)).))).... ( -25.82) >DroSim_CAF1 2442 108 - 1 AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUGCGCUACGUGCACCUGAAAGGGACAAAAGGGUU---------GACGCAGC---CUGCAUGCAAAGCAGCCAGAG ..........(((..........(((((.........)))))...(((....(((((.(((....)))......(((((---------(...))))---)))))))....)))))).... ( -28.20) >DroEre_CAF1 2095 117 - 1 AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUGCGCUACGUGCACCUGAAAGGGACAAAAGGGUUGACGCAGUUGACGCAGC---CUGCAUGCAAAGCGGCCAGAG ..........((((.........(((((.........)))))......(((.(((((.(((....)))......((((((.(((....).))))))---)))))))...))))))).... ( -35.30) >DroYak_CAF1 2369 113 - 1 AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUGCGCUACGUGCACCUGAAAGGGACAAAAGGGUUG-------UGACGCAUUGUGAUGCAUGCAAAGCAGCCAGAG .......((((............(((((.........)))))..((((((...))))))))))............(((((-------(...((((.(.....)))))...)))))).... ( -26.80) >consensus AUAAAGGUCAGGCCAAUAAAAACCUAUCAAAACUGAAGAUAGAUGUGCGCUACGUGCACCUGAAAGGGACAAAAGGGUU_________GACGCAGC___CUGCAUGCAAAGCAGCCAGAG ..........(((..........(((((.........)))))......(((...(((((((....))).......................((((....)))).)))).))).))).... (-22.56 = -22.64 + 0.08)

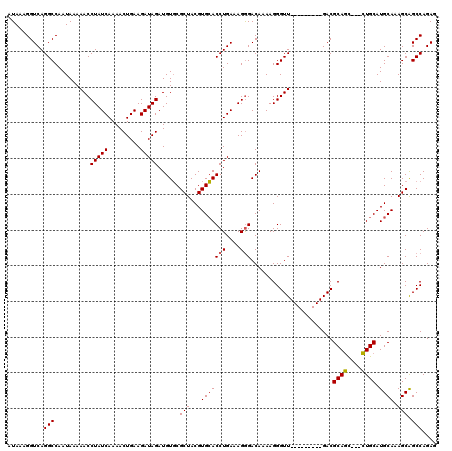
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:54 2006