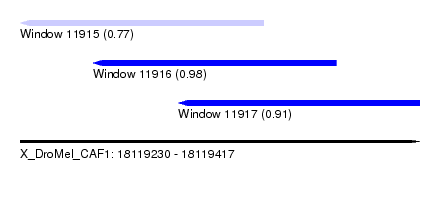
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,119,230 – 18,119,417 |
| Length | 187 |
| Max. P | 0.980142 |

| Location | 18,119,230 – 18,119,344 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -31.34 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18119230 114 - 22224390 GCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGAGUCGGAGUAGGAGUAGCUGAUGCAGUUGGAGAUG------ ((((((((((..((((.....))))))))))))))..((((((..((.((((......((((.((..((.....))..)).))))......))))((....)).))..))))))------ ( -37.30) >DroSec_CAF1 1844 114 - 1 GCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGAGUCGGAGUAGGAGUCACAGAUGGAGAUGGAGGUG------ ((((((((((..((((.....))))))))))))))(((((((.((((.......((((......))))..((((.......)))).......)))).)))))))..........------ ( -37.20) >DroSim_CAF1 2115 120 - 1 GCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGAGUCGGAGUAGGAGUCACAGAUGGAGAUGGAGGUGGAGAUG ((((((((((..((((.....))))))))))))))((((((((..(((........)))..))((.(((.(((.....((.((.......)).)).....))).))).)))))))).... ( -38.40) >DroEre_CAF1 1769 113 - 1 GAGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGAGUCGGAGUCAGAGUCG-GGAUCGGGAUCGGGAUG------ ..((((((((..((((.....)))))))))))).(((.(((((..(((........)))..)).......((((....((.((.......)).)).-...))))))).)))...------ ( -32.30) >consensus GCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGAGUCGGAGUAGGAGUCACAGAUGGAGAUGGAGAUG______ ((((((((((..((((.....))))))))))))))(((((((.((((.......((((......))))..((((.......)))).......)))).)))))))................ (-31.34 = -32.02 + 0.69)



| Location | 18,119,264 – 18,119,378 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.74 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -30.25 |
| Energy contribution | -32.38 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18119264 114 - 22224390 AUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGA .(((((..(((.(((..---...((((((((((....((((((((((..((((.....))))))))))))))))))))))))(((........))).......))).)))..))))) ( -42.00) >DroSec_CAF1 1878 114 - 1 AUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGA .(((((..(((.(((..---...((((((((((....((((((((((..((((.....))))))))))))))))))))))))(((........))).......))).)))..))))) ( -42.00) >DroSim_CAF1 2155 114 - 1 AUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGA .(((((..(((.(((..---...((((((((((....((((((((((..((((.....))))))))))))))))))))))))(((........))).......))).)))..))))) ( -42.00) >DroYak_CAF1 2068 101 - 1 AUCUUAACUGGAGACGGAGA---------------UGGAGAAUGGGAAUGGGAAUGCAUCUCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGG- ..(((...(((.(((.((((---------------(((((....(((..((((.....))))..)))....)))))))((..(((........)))..)))).))).)))..))).- ( -27.60) >consensus AUCUUUACUGGAGACGC___AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAUCUUUGGCAUAUUAAAUUGUUGAAUCAGUCACCGAGAAGGA .(((((..(((.(((............(((((....(((((((((((..((((.....))))))))))))))))))))((..(((........)))..))...))).)))..))))) (-30.25 = -32.38 + 2.12)

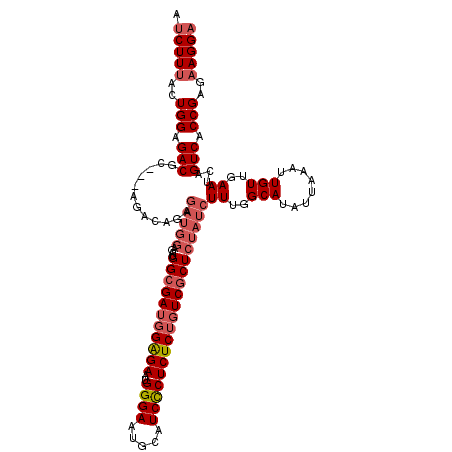
| Location | 18,119,304 – 18,119,417 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -26.85 |
| Energy contribution | -28.10 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18119304 113 - 22224390 GCCGCAGGGAGAGCACACAAUAAAAUCAGACC-GAUAACCAUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAU .((....))(((((..............(.((-.((..((((((((.(((......)---))..))))))))..)))))((((((((..((((.....))))))))))))))))).. ( -36.70) >DroSec_CAF1 1918 113 - 1 GCCACAGGGAGAGCACACAAUAAAAUCAGACC-GAUAACCAUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAU .((....))(((((..............(.((-.((..((((((((.(((......)---))..))))))))..)))))((((((((..((((.....))))))))))))))))).. ( -36.50) >DroSim_CAF1 2195 113 - 1 GCCGCAGGGAGAGCACACAAUAAAAUCAGACC-GAUAACCAUCUUUACUGGAGACGC---AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAU .((....))(((((..............(.((-.((..((((((((.(((......)---))..))))))))..)))))((((((((..((((.....))))))))))))))))).. ( -36.70) >DroYak_CAF1 2107 102 - 1 GCCGCAGGGAGAGCACACAAUAAAAUCAGUCCUGAUAACCAUCUUAACUGGAGACGGAGA---------------UGGAGAAUGGGAAUGGGAAUGCAUCUCUCUCUGUCGCUCUAU ((.((((((((((....((.(....(((.(((((.(..((((((...(((....))))))---------------)))..).))))).))).).))...)))))))))).))..... ( -35.20) >consensus GCCGCAGGGAGAGCACACAAUAAAAUCAGACC_GAUAACCAUCUUUACUGGAGACGC___AGACAGAGAUGGAGAUGGCGAUGGAGAAUGGGAAUGCAUCCCUCUCUGUCGCUCUAU .((....))(((((........................(((((((....(....)........((....))))))))).((((((((..((((.....))))))))))))))))).. (-26.85 = -28.10 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:53 2006