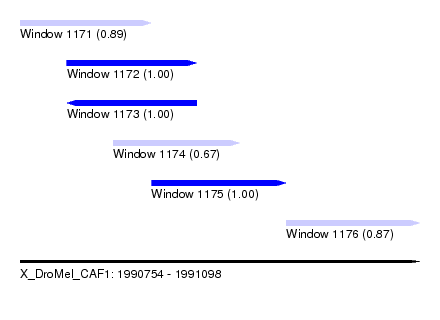
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,990,754 – 1,991,098 |
| Length | 344 |
| Max. P | 0.999981 |

| Location | 1,990,754 – 1,990,867 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990754 113 + 22224390 CAAGAGCAUAAAAAGCAUGAGUGCAAAAAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAA .....((.......(((....)))...............((.(((((...(((((.(((((((........)).)))))..))))).....))))).)).....)).-------...... ( -22.90) >DroSec_CAF1 22758 113 + 1 CAAGAGCAUAAAAAGCAUGAGUGCAAAAAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAA .....((.......(((....)))...............((.(((((...(((((.(((((((........)).)))))..))))).....))))).)).....)).-------...... ( -22.90) >DroSim_CAF1 17204 113 + 1 CAAGAGCAUAAAAAGCAUGAGUGCAAAAAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAA .....((.......(((....)))...............((.(((((...(((((.(((((((........)).)))))..))))).....))))).)).....)).-------...... ( -22.90) >DroEre_CAF1 12479 120 + 1 CAAGAGCAUAAAAAGCAUGAGUGCAAAGAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUAAACCGUAAAAGAA .....((.......(((....)))...............((.(((((...(((((.(((((((........)).)))))..))))).....))))).)).....)).............. ( -22.90) >DroYak_CAF1 23274 120 + 1 CAAAAGCAUAAAAAGCAUGAGUGCAAAGAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUAAACUGUUAAAGAA .....((((...........))))......(((((.....(.(((((...(((((.(((((((........)).)))))..))))).....))))).)(((...)))..)))))...... ( -25.00) >consensus CAAGAGCAUAAAAAGCAUGAGUGCAAAAAAGACAGAAAAAGGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA_______AAAGAA .....((.......(((....)))...............((.(((((...(((((.(((((((........)).)))))..))))).....))))).)).....)).............. (-22.90 = -22.90 + 0.00)

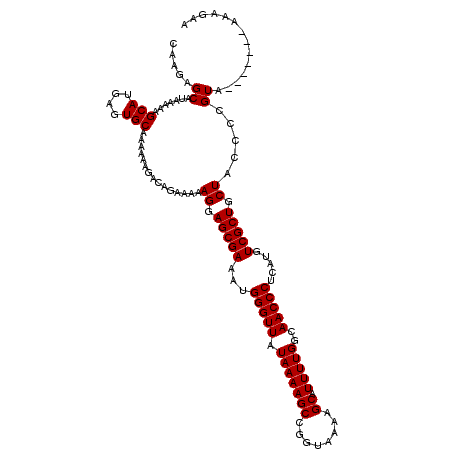
| Location | 1,990,794 – 1,990,906 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -37.57 |
| Energy contribution | -38.17 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.26 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990794 112 + 22224390 GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAAUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).((((((((((-------........-....))))))))))(((.((((......)))).))). ( -36.80) >DroSec_CAF1 22798 112 + 1 GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).((((((((((-------........-....))))))))))((((((((......)))))))). ( -40.40) >DroSim_CAF1 17244 112 + 1 GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).((((((((((-------........-....))))))))))((((((((......)))))))). ( -40.40) >DroEre_CAF1 12519 120 + 1 GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUAAACCGUAAAAGAAAGGGGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).(((((((.....((.(.......).))......)))))))((((((((......)))))))). ( -40.70) >DroYak_CAF1 23314 119 + 1 GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUAAACUGUUAAAGAAAG-GGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).(((((((.....((.((.....)).-)).....)))))))((((((((......)))))))). ( -39.60) >consensus GGAGCGAAAUGGGUUAUAAAAGCCGGUAAAAGCAUUUUGGCAACCCUCAUGUCGCUGCUACCCCGUA_______AAAGAAAG_GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACU ..(((((...(((((.(((((((........)).)))))..))))).....))))).((((((((((....................))))))))))((((((((......)))))))). (-37.57 = -38.17 + 0.60)

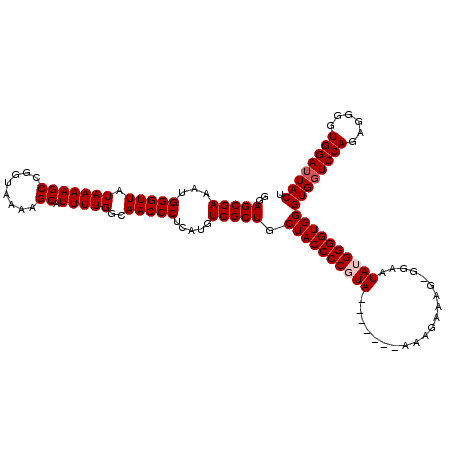
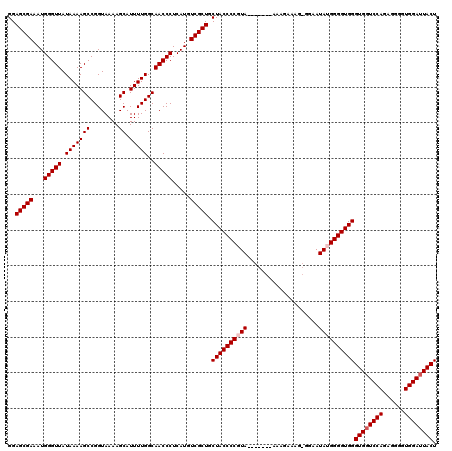
| Location | 1,990,794 – 1,990,906 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990794 112 - 22224390 AGUAUUCCACCCCUCUGGACCACCCACCCCAUAUUCC-CUUUCUUU-------UACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((.((....-........-------)).)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. ( -25.50) >DroSec_CAF1 22798 112 - 1 AGUAAUCCACCCCUCUGGACCACCCACCCCAUAUUCC-CUUUCUUU-------UACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((.((....-........-------)).)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. ( -25.50) >DroSim_CAF1 17244 112 - 1 AGUAAUCCACCCCUCUGGACCACCCACCCCAUAUUCC-CUUUCUUU-------UACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((.((....-........-------)).)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. ( -25.50) >DroEre_CAF1 12519 120 - 1 AGUAAUCCACCCCUCUGGACCACCCACCCCUUAUUCCCCUUUCUUUUACGGUUUACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((.((..((............))..)).)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. ( -28.60) >DroYak_CAF1 23314 119 - 1 AGUAAUCCACCCCUCUGGACCACCCACCCCUUAUUCC-CUUUCUUUAACAGUUUACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((.((..(.-.((.....))..)..)).)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. ( -25.60) >consensus AGUAAUCCACCCCUCUGGACCACCCACCCCAUAUUCC_CUUUCUUU_______UACGGGGUAGCAGCGACAUGAGGGUUGCCAAAAUGCUUUUACCGGCUUUUAUAACCCAUUUCGCUCC .((..((((......))))..))..(((((..........................)))))...(((((.((..((((((..((((.(((......))))))).)))))))).))))).. (-24.83 = -24.83 + 0.00)

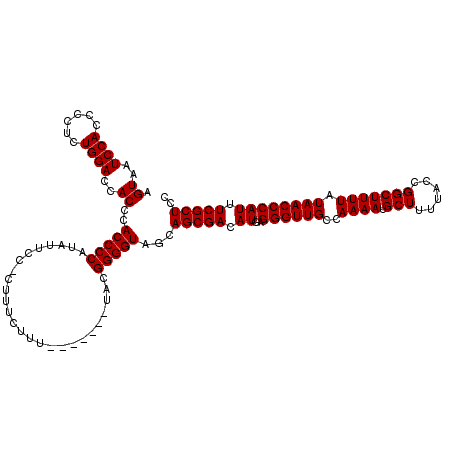
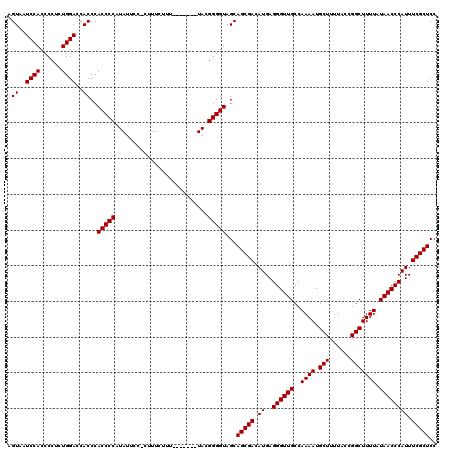
| Location | 1,990,834 – 1,990,943 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -33.53 |
| Energy contribution | -34.13 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990834 109 + 22224390 CAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAAUACUCGCCACAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUG ((.(((((.((..((..((((((((((-------........-....)))))))).))..)).))))))).))...((((((((((...)))---)))))))((((..(....)..)))) ( -42.80) >DroSec_CAF1 22838 109 + 1 CAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCAAAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUG ((.(((((.((..((..((((((((((-------........-....)))))))).))..)).))))))).))...(((((((((.....))---)))))))((((..(....)..)))) ( -40.90) >DroSim_CAF1 17284 109 + 1 CAACCCUCAUGUCGCUGCUACCCCGUA-------AAAGAAAG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCAAAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUG ((.(((((.((..((..((((((((((-------........-....)))))))).))..)).))))))).))...(((((((((.....))---)))))))((((..(....)..)))) ( -40.90) >DroEre_CAF1 12559 117 + 1 CAACCCUCAUGUCGCUGCUACCCCGUAAACCGUAAAAGAAAGGGGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACUGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUG ((.(((((.((..((..(((((((.....((.(.......).))......))))).))..)).))))))).))...((((((((((...)))---)))))))((((..(....)..)))) ( -43.00) >DroYak_CAF1 23354 119 + 1 CAACCCUCAUGUCGCUGCUACCCCGUAAACUGUUAAAGAAAG-GGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACUGGGUGGUGGUGGGUGCGCGUGUGCAAGUGUGUG ((.(((((.((..((..(((((((.....((.((.....)).-)).....))))).))..)).))))))).))...(((((((((((....)))))))))))((((..(....)..)))) ( -44.80) >consensus CAACCCUCAUGUCGCUGCUACCCCGUA_______AAAGAAAG_GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACAGGGUG___GUGGGUGCGCGUGUGCAAGUGUGUG ..((((.(.........((((((((((....................))))))))))((((((((......)))))))).((((....))))...).))))((((........))))... (-33.53 = -34.13 + 0.60)

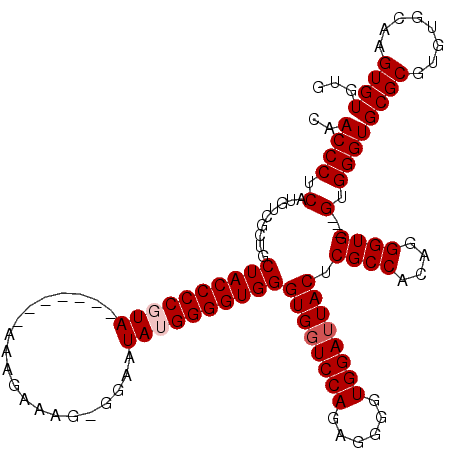
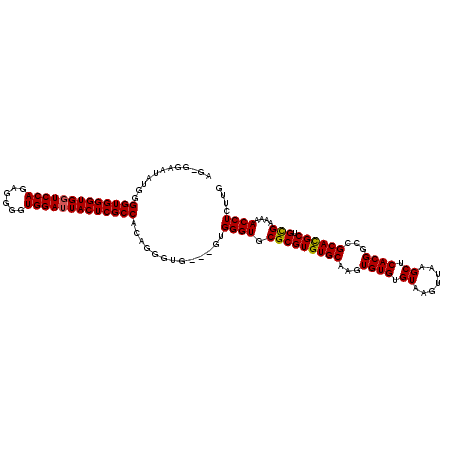
| Location | 1,990,867 – 1,990,983 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -41.06 |
| Energy contribution | -40.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990867 116 + 22224390 AG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAAUACUCGCCACAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUAAGCUCACGGCCGCACGCUGCGAAAAACCUCUUG ..-.........((((((((.((((......)))).)))))))).(((((.(---((...((((((((((...(((.((.........)).)))...))))))).)))....)))))))) ( -43.30) >DroSec_CAF1 22871 116 + 1 AG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCAAAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUAAGCUCACGGCCGCACGCUGCGAAAAACCUCUUG ..-.........(((((((((((((......)))))))))))))...(((.(---((...((((((((((...(((.((.........)).)))...))))))).)))....))).))). ( -45.20) >DroSim_CAF1 17317 116 + 1 AG-GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCAAAGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUAAGCUCACGGCCGCAUGCUGCGAAAAACCUCUUG ..-.........(((((((((((((......)))))))))))))...(((.(---((...((((((((((...(((.((.........)).)))...))))))).)))....))).))). ( -43.40) >DroEre_CAF1 12599 117 + 1 AGGGGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACUGGGUG---GUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUGAGCUCACGGCCGCACGCUGUGAAAAACCUCUUG (((((....((.(((((((((((((......))))))))))))).)).(((.---((((((..((.((..((....))..)).))...)))))).)))...............))))).. ( -45.30) >DroYak_CAF1 23394 119 + 1 AG-GGAAUAAGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACUGGGUGGUGGUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUGAGCUCACGGCCGCACGCUGCGAAAAACCUCUUG ..-....((((((((((((((((((......))))))))))((((((....))))))...((((((((((...(((.((.........)).)))...))))))).)))....)))))))) ( -47.70) >consensus AG_GGAAUAUGGGGUGGGUGGUCCAGAGGGGUGGAUUACUCGCCACAGGGUG___GUGGGUGCGCGUGUGCAAGUGUGUGUAAGUUAAGCUCACGGCCGCACGCUGCGAAAAACCUCUUG ............(((((((((((((......))))))))))))).............((((.(((((((((...((((.((.......)).))))...)))))).)))....)))).... (-41.06 = -40.94 + -0.12)

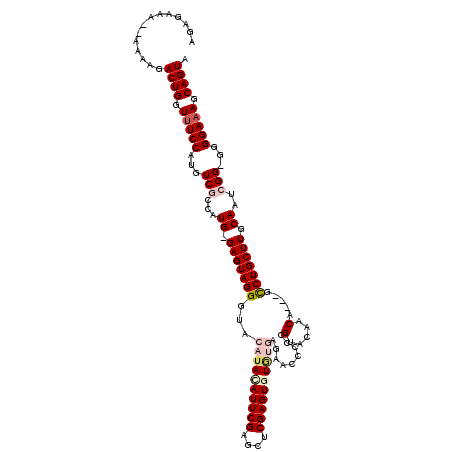
| Location | 1,990,983 – 1,991,098 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -28.54 |
| Energy contribution | -30.10 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1990983 115 + 22224390 AGAGAAAAAAAAAGACUGGUUUCCAUGUCGCCAUG-GAGUAGGGUACAUACAUUCGAGCUCGAGUGUGUGAGAACCCCCGUAAAACA---GCCUGCUUGCAAUCGG-GGGGAAAGCAGUA ..............((((..(((((((....))))-))).......((((((((((....))))))))))....((((((......(---(....))......)))-))).....)))). ( -38.10) >DroSec_CAF1 22987 113 + 1 AGAGAAA--AAAAGACUGGUUUCCAUGUCGCCAUG-GAGUAGGGUACAUACAUUCGAGCUCGAGUAUGUGAGAACCCCCGUACAACA---GCCUGCUUGCAAUCGG-GGGGAAAGCAGUA .......--.....((((..(((((((....))))-))).....(((((((..(((....))))))))))....((((((......(---(....))......)))-))).....)))). ( -32.80) >DroSim_CAF1 17433 113 + 1 AGAGAAA--AAAAGACUGGUUUCCAUGUCGCCAUG-GAGUAGGGUACAUACAUUCGAGCUCGAGUAUGUGAGAACCCCCGUACAACA---GCCUGCUUGCAAUCGG-GGGGAAAGCAGUA .......--.....((((..(((((((....))))-))).....(((((((..(((....))))))))))....((((((......(---(....))......)))-))).....)))). ( -32.80) >DroEre_CAF1 12716 100 + 1 AGAGAGA---AAAGACUGGUUUCCA-GUCGCCAUG-GAGUAGGGUACAUAUAUUCGAGCUCGAGUGUA---------CCGUACAACA---GUCUGCUUGCAACGGG---GGAAAGCAGUA .......---....((((.(((((.-((.((....-(((((((((((.((((((((....))))))))---------..))))....---.))))))))).))...---))))).)))). ( -31.60) >DroYak_CAF1 23513 114 + 1 AGAGAAAAAAAAAGACUGGUAUCCA-GUCGCCAUGGGAGUAGGGU----AUAUUCGAGCUCGAGUGUGCGAGAACC-CCGUACAACAACCGUCUGCUUGCAACGGGGGGGGAAAGCAGUA .............((((((...)))-)))((.(((((.((...((----(((((((....)))))))))....)))-)))).(..(..((((.((....))))))..)..)...)).... ( -36.20) >consensus AGAGAAA__AAAAGACUGGUUUCCAUGUCGCCAUG_GAGUAGGGUACAUACAUUCGAGCUCGAGUGUGUGAGAACCCCCGUACAACA___GCCUGCUUGCAAUCGG_GGGGAAAGCAGUA ..............((((.(((((...(((...((.(((((((...((((((((((....)))))))))).........(.....).....))))))).))..)))...))))).)))). (-28.54 = -30.10 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:17 2006