
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,110,895 – 18,111,004 |
| Length | 109 |
| Max. P | 0.975258 |

| Location | 18,110,895 – 18,111,004 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -34.97 |
| Energy contribution | -35.75 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
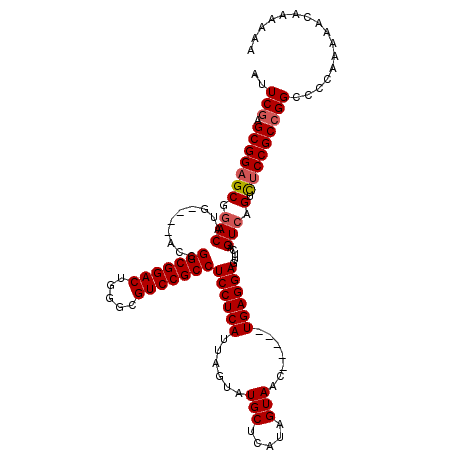
>X_DroMel_CAF1 18110895 109 + 22224390 AUUCGAGCGGAGCGAACAUG----ACCGGCGGACUGGGCGUCCGCCUCCUCAUUAGUAUGCUCAUAGUAAC-----UGAGGAGUUCAGUCAGUCUCCGCCGGCCACAAAAACAAAAAA ..(((.(((((((..((.((----(.((((((((.....)))))))(((((((((.(((....))).))).-----))))))).)))))..).)))))))))................ ( -42.30) >DroSec_CAF1 19361 109 + 1 AUUCGAGCGGAGCGGACAUG----ACCGGCGGACUGGGCGUCCGCCUCCUCAUUAGUAUGCUCAUAGUAAC-----UGAGGAGUUCAGUCAGUCUCCGCCGGACCCAAAAACAAAAAA .((((.(((((((.(((.((----(.((((((((.....)))))))(((((((((.(((....))).))).-----))))))).)))))).).))))))))))............... ( -46.30) >DroYak_CAF1 19201 115 + 1 CUUCAAGCGGAGCGGACCGGACCG-UCGGCGGACUGGGCGUCCGCCUCCUCAUUAGUAUGCUCAUAGUAACUGAACUGAGGAGUUCAGUCAGUUCCCGCCGGCCCGAAAAAGAAAU-- ......(((((((.(((.((((..-..(((((((.....)))))))(((((((((((.(((.....))))))))..)))))))))).))).))).)))).................-- ( -43.50) >consensus AUUCGAGCGGAGCGGACAUG____ACCGGCGGACUGGGCGUCCGCCUCCUCAUUAGUAUGCUCAUAGUAAC_____UGAGGAGUUCAGUCAGUCUCCGCCGGCCCCAAAAACAAAAAA ..(((.(((((((.(((..........(((((((.....)))))))((((((......(((.....))).......)))))).....))).).)))))))))................ (-34.97 = -35.75 + 0.78)

| Location | 18,110,895 – 18,111,004 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -47.57 |
| Consensus MFE | -37.16 |
| Energy contribution | -36.83 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
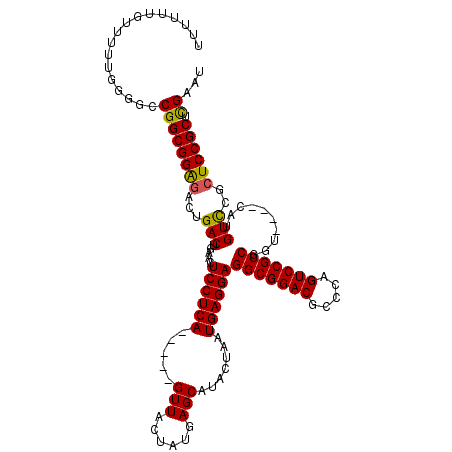
>X_DroMel_CAF1 18110895 109 - 22224390 UUUUUUGUUUUUGUGGCCGGCGGAGACUGACUGAACUCCUCA-----GUUACUAUGAGCAUACUAAUGAGGAGGCGGACGCCCAGUCCGCCGGU----CAUGUUCGCUCCGCUCGAAU ......((((..((((..((((((.(.((((((..(((((((-----.(((.(((....))).))))))))))((((((.....))))))))))----))).))))))))))..)))) ( -44.50) >DroSec_CAF1 19361 109 - 1 UUUUUUGUUUUUGGGUCCGGCGGAGACUGACUGAACUCCUCA-----GUUACUAUGAGCAUACUAAUGAGGAGGCGGACGCCCAGUCCGCCGGU----CAUGUCCGCUCCGCUCGAAU .........(((((((..((((((.(.((((((..(((((((-----.(((.(((....))).))))))))))((((((.....))))))))))----))).))))))..))))))). ( -48.60) >DroYak_CAF1 19201 115 - 1 --AUUUCUUUUUCGGGCCGGCGGGAACUGACUGAACUCCUCAGUUCAGUUACUAUGAGCAUACUAAUGAGGAGGCGGACGCCCAGUCCGCCGA-CGGUCCGGUCCGCUCCGCUUGAAG --........(((((((.((((((....(((((...(((((((((((.......))))).......))))))(((((((.....)))))))..-)))))...))))).).))))))). ( -49.61) >consensus UUUUUUGUUUUUGGGGCCGGCGGAGACUGACUGAACUCCUCA_____GUUACUAUGAGCAUACUAAUGAGGAGGCGGACGCCCAGUCCGCCGGU____CAUGUCCGCUCCGCUCGAAU .................((((((((...(((.....((((((.....(((......))).......))))))(((((((.....)))))))..........)))..)))))).))... (-37.16 = -36.83 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:42 2006