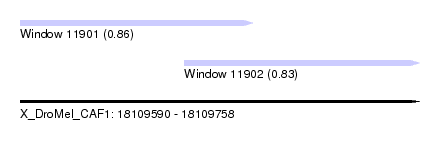
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,109,590 – 18,109,758 |
| Length | 168 |
| Max. P | 0.862887 |

| Location | 18,109,590 – 18,109,688 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.29 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18109590 98 + 22224390 GGAGUGGCAGUGGCAGUGAGCGGUACACGGUACAUGGUACGCGGCAAAGCGUGAACCGUAAACGGUAAA-------CGACAAAACGGAGACGUCGCCACUUUGGC ((((((((.((.((.((..(((((.(((.((((...))))((......))))).)))))..)).))..)-------)......(((....))).))))))))... ( -35.20) >DroSec_CAF1 18043 97 + 1 GGAGUGGCAGUGGCAGUGAGCGGUACACGGUACAUGGAACGCGGCAAGGCGUGAACCGUAAACGGUAAA-------CGACAA-ACGGCGACGUCGCCACUUUGGC ((((((((.((.((.((.....((((...)))).....)))).))..(((((...((((...((.....-------))....-))))..)))))))))))))... ( -32.30) >DroEre_CAF1 15926 87 + 1 ---GCCGCAGUGGGAGUGAGCGGUACGCGGU--------------GAAGCGUGAACCGCAAACGGUAAAUAUUAAACGAGAA-ACGGAGACGUCGCCGCUUUGAC ---..((.(((((..((..(((((.((((..--------------....)))).)))))..))...................-(((....)))..))))).)).. ( -27.20) >consensus GGAGUGGCAGUGGCAGUGAGCGGUACACGGUACAUGG_ACGCGGCAAAGCGUGAACCGUAAACGGUAAA_______CGACAA_ACGGAGACGUCGCCACUUUGGC ((((((((.......((..(((((.((((.(................).)))).)))))..))....................(((....))).))))))))... (-26.06 = -26.29 + 0.23)

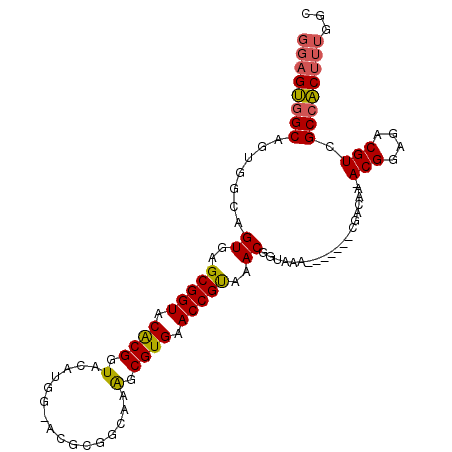
| Location | 18,109,659 – 18,109,758 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18109659 99 + 22224390 ----------CGACAAAACGGAGACGUCGCCACUUUGG-CCAUGAGUAACGAUGAGUGCUCAAUUACUUUUCGAUUGAUCAUGGUUUUGGCGCGCGGUGAAAUUUAGACC---------- ----------((.(...(((....)))(((((....((-((((((.((((((.(((((......))))).))).))).)))))))).))))).)))..............---------- ( -29.80) >DroSec_CAF1 18112 107 + 1 ----------CGACAA-ACGGCGACGUCGCCACUUUGG-CCAUGAGUAACGAUGAGUGCUCAAUUACUUUUCGAUUGAUCAUGGUUUUGGCGCGCGGUGAAAUUUAGACCGCUUUUCAG- ----------......-(((....)))(((((....((-((((((.((((((.(((((......))))).))).))).)))))))).))))).(((((.........))))).......- ( -34.70) >DroEre_CAF1 15985 108 + 1 ----------CGAGAA-ACGGAGACGUCGCCGCUUUGA-CCAUGAGUAACGACGAGUGCUCAAUUACUUUUCGAUUGAUCAUGGUUUUGGCGCGCGGUGAAAUUUAGACCGCUAUUCAGA ----------...(((-(((....))).((.(((..((-((((((.((((((.(((((......))))).))).))).))))))))..)))))(((((.........)))))..)))... ( -38.20) >DroYak_CAF1 17885 119 + 1 AAGCAACAAGCGACAA-ACGGCGACGUCGCCACUUAGUUCCAUGAGUAACGAUGAGUGCUCAAUUACUUUUCGAUUGAUCAUGGUUUUGGCGCGCGGUGAAAUUUAGACCGCUUUUCAGA .........(((.(((-(.((((....))))........((((((.((((((.(((((......))))).))).))).)))))).)))).)))(((((.........)))))........ ( -36.30) >consensus __________CGACAA_ACGGAGACGUCGCCACUUUGG_CCAUGAGUAACGAUGAGUGCUCAAUUACUUUUCGAUUGAUCAUGGUUUUGGCGCGCGGUGAAAUUUAGACCGCUUUUCAG_ .................(((....)))(((((.......((((((.((((((.(((((......))))).))).))).))))))...))))).(((((.........)))))........ (-28.76 = -29.08 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:39 2006