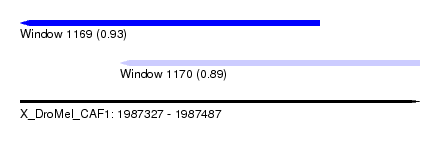
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,987,327 – 1,987,487 |
| Length | 160 |
| Max. P | 0.926085 |

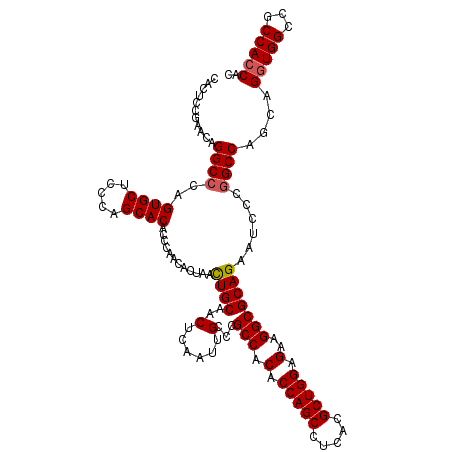
| Location | 1,987,327 – 1,987,447 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -33.91 |
| Energy contribution | -34.23 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1987327 120 - 22224390 CACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGGAGAAGGCGCAGAAUCCCGGCCAGCAGGUGGCCGCCACCAC ...........((((..((((.....))))............((((..(......)...(((.(.(((((.....))))).)..)))))))......))))....(((((...))))).. ( -37.50) >DroSec_CAF1 20865 119 - 1 CACUCCGAACAGGC-CAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGGAGAAGGCGCAGAAUCCCGGCCAGCAGGUGGCCGCCAGCAC ...........(((-(.((((.....))))............((((..(......)...(((.(.(((((.....))))).)..)))))))......)))).((.(((....))).)).. ( -36.90) >DroSim_CAF1 14630 120 - 1 CACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGGAGAAGGCGCAGAAUCCCGGCCAGCAGGUGGCCGCCACCAC ...........((((..((((.....))))............((((..(......)...(((.(.(((((.....))))).)..)))))))......))))....(((((...))))).. ( -37.50) >DroEre_CAF1 10460 120 - 1 CACUCCAAACAGGCCCAGUGCACCCAGCACACCCAACACUAAUUGCAACUCCAUUGCCCGCCACACCAGCCUCACGCUGGAGAAGGCGCAGAAUCCCGGCCAGCAGGUGGCCACCACCAC ...........((((..((((.....))))..............((((.....)))).((((.(.(((((.....))))).)..)))).........))))....(((((...))))).. ( -35.60) >consensus CACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGGAGAAGGCGCAGAAUCCCGGCCAGCAGGUGGCCGCCACCAC ...........((((..((((.....))))............((((..(......)...(((.(.(((((.....))))).)..)))))))......))))....(((((...))))).. (-33.91 = -34.23 + 0.31)

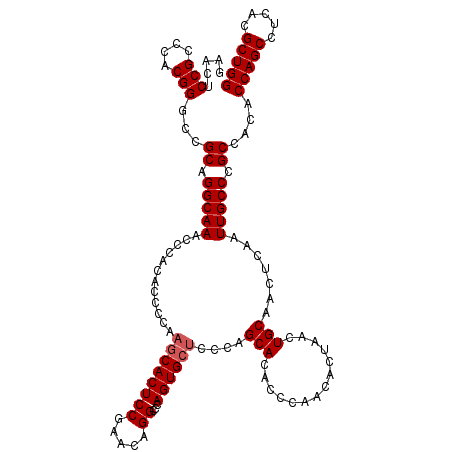
| Location | 1,987,367 – 1,987,487 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -25.51 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1987367 120 - 22224390 GAACUCCGCCCACGGGCCGCAGGCAAACCCACACCCCAAGCACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGG .......(((....))).((.(((((............((((((((.....))...))))))....(((..............))).......))))).))....(((((.....))))) ( -30.34) >DroSec_CAF1 20905 117 - 1 GAACUCCGC-CACGGGCCGCAGGCAAACCCACACC-CAAGCACUCCGAACAGGC-CAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGG .......((-(...))).((.(((((.........-..((((((((.....)).-.))))))....(((..............))).......))))).))....(((((.....))))) ( -27.74) >DroSim_CAF1 14670 120 - 1 GAACUCCGCCCACGGGCCGCAGGCAAACCCACACCCCAAGCACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGG .......(((....))).((.(((((............((((((((.....))...))))))....(((..............))).......))))).))....(((((.....))))) ( -30.34) >DroEre_CAF1 10500 120 - 1 GAACUCCGCCCACGGUCCGCAGGCAAACCCACACCCCAAACACUCCAAACAGGCCCAGUGCACCCAGCACACCCAACACUAAUUGCAACUCCAUUGCCCGCCACACCAGCCUCACGCUGG .....(((....)))...((.(((((..................((.....))....((((.....)))).......................))))).))....(((((.....))))) ( -23.60) >consensus GAACUCCGCCCACGGGCCGCAGGCAAACCCACACCCCAAGCACUCCGAACAGGCCCAGUGCUCCCAGCACACCCAACACUAACUGCAACUCAAUUGCCCGCCACACCAGCCUCACGCUGG .....(((....)))...((.(((((............((((((((.....))...))))))....(((..............))).......))))).))....(((((.....))))) (-25.51 = -26.02 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:11 2006