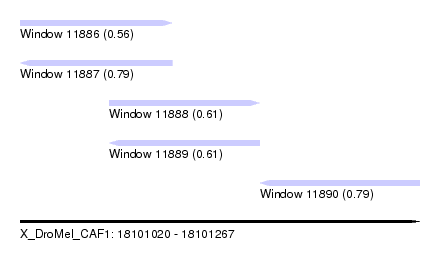
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,101,020 – 18,101,267 |
| Length | 247 |
| Max. P | 0.791869 |

| Location | 18,101,020 – 18,101,114 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -29.13 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18101020 94 + 22224390 ----CGACCGGCUGUUAUGCAAAUGCGACCGCC-------------------GU--GGGGAGACGGAGACAUCGCCAUCUUGGCAGGCCGAUGGCUGAUCUC-UCGCCAUCUUGGUCGCC ----((((((((.(((..((....))))).)))-------------------((--((.((((.(....)(((((((((..((....)))))))).))).))-)).))))...))))).. ( -40.10) >DroSec_CAF1 9648 94 + 1 ----CGACCGGCUGUUAUGCAAAUGCGACCGCC-------------------GU--GGGGAGACGGAGACAUCGCCAUCUUGGCAGGCCGAUGGCUGAUCUC-UCGCCAUCUUGGUCGCC ----((((((((.(((..((....))))).)))-------------------((--((.((((.(....)(((((((((..((....)))))))).))).))-)).))))...))))).. ( -40.10) >DroSim_CAF1 9722 94 + 1 ----CGACCGGCUGUUAUGCAAAUGCGACCGCC-------------------GU--GGGGAGACGGAGACAUCGCCAUCUUGGCAGGCCGAUGGCUGAUCUC-UCGCCAUCUUGGUCGCC ----((((((((.(((..((....))))).)))-------------------((--((.((((.(....)(((((((((..((....)))))))).))).))-)).))))...))))).. ( -40.10) >DroEre_CAF1 7100 94 + 1 ----CGACCGGCUGUUAUGCAAAUGCGACCGCC-------------------GU--GGGUAGACGGAGACAUCGCCAUCUUGGCAGGCCGAUGGCUGAUCUC-UCGCCAUCUUGGUCGCC ----(((((((((....(((....)))...(((-------------------(.--.(((.((.(....).)))))....)))).))))((((((.((....-))))))))..))))).. ( -38.30) >DroYak_CAF1 8777 100 + 1 ACAGCGACCGGCUGUUAUGCAAAUGCGACCGCC-------------------GUGUGGGGAGACGGAGACAUCGCCAUCUUGGCUGGCCGAUGGCUGAUCUC-UCGCCAUCUUGGUCGCC ...((((((((((....(((....)))...(((-------------------(.((((.((...(....).)).))))..)))).))))((((((.((....-))))))))..)))))). ( -44.70) >DroAna_CAF1 12054 109 + 1 ----GGGUGGGCUGUUAUGCAAAUAUAUUCUCCAAAGAGGGGGGGGGGAGGGGA--GGAGAAAAGGAGACAUCGCCAUCUUGGCAGG-AGAUGGCUGAUCUCUUCGCCAUCUUUGG---- ----(((((.((......)).....))))).(((((((.((.(((((((.....--........(....)...((((((((.....)-)))))))...))))))).)).)))))))---- ( -38.50) >consensus ____CGACCGGCUGUUAUGCAAAUGCGACCGCC___________________GU__GGGGAGACGGAGACAUCGCCAUCUUGGCAGGCCGAUGGCUGAUCUC_UCGCCAUCUUGGUCGCC ..........((......))....(((((((((............................((.(....).))(((.....))).))).((((((.((.....))))))))..)))))). (-29.13 = -29.72 + 0.59)

| Location | 18,101,020 – 18,101,114 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -30.15 |
| Energy contribution | -30.79 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18101020 94 - 22224390 GGCGACCAAGAUGGCGA-GAGAUCAGCCAUCGGCCUGCCAAGAUGGCGAUGUCUCCGUCUCCCC--AC-------------------GGCGGUCGCAUUUGCAUAACAGCCGGUCG---- ..(((((..((((((((-....)).))))))(((..(((.....)))(((((..(((((.....--..-------------------)))))..))))).........))))))))---- ( -39.30) >DroSec_CAF1 9648 94 - 1 GGCGACCAAGAUGGCGA-GAGAUCAGCCAUCGGCCUGCCAAGAUGGCGAUGUCUCCGUCUCCCC--AC-------------------GGCGGUCGCAUUUGCAUAACAGCCGGUCG---- ..(((((..((((((((-....)).))))))(((..(((.....)))(((((..(((((.....--..-------------------)))))..))))).........))))))))---- ( -39.30) >DroSim_CAF1 9722 94 - 1 GGCGACCAAGAUGGCGA-GAGAUCAGCCAUCGGCCUGCCAAGAUGGCGAUGUCUCCGUCUCCCC--AC-------------------GGCGGUCGCAUUUGCAUAACAGCCGGUCG---- ..(((((..((((((((-....)).))))))(((..(((.....)))(((((..(((((.....--..-------------------)))))..))))).........))))))))---- ( -39.30) >DroEre_CAF1 7100 94 - 1 GGCGACCAAGAUGGCGA-GAGAUCAGCCAUCGGCCUGCCAAGAUGGCGAUGUCUCCGUCUACCC--AC-------------------GGCGGUCGCAUUUGCAUAACAGCCGGUCG---- ..(((((..((((((((-....)).))))))(((..(((.....)))(((((..(((((.....--..-------------------)))))..))))).........))))))))---- ( -39.30) >DroYak_CAF1 8777 100 - 1 GGCGACCAAGAUGGCGA-GAGAUCAGCCAUCGGCCAGCCAAGAUGGCGAUGUCUCCGUCUCCCCACAC-------------------GGCGGUCGCAUUUGCAUAACAGCCGGUCGCUGU (((((((..((((((((-....)).))))))(((..(((.....)))(((((..(((((.........-------------------)))))..))))).........)))))))))).. ( -44.50) >DroAna_CAF1 12054 109 - 1 ----CCAAAGAUGGCGAAGAGAUCAGCCAUCU-CCUGCCAAGAUGGCGAUGUCUCCUUUUCUCC--UCCCCUCCCCCCCCCCUCUUUGGAGAAUAUAUUUGCAUAACAGCCCACCC---- ----........(((...(((((..(((((((-.......)))))))...)))))...((((((--.....................))))))...............))).....---- ( -27.00) >consensus GGCGACCAAGAUGGCGA_GAGAUCAGCCAUCGGCCUGCCAAGAUGGCGAUGUCUCCGUCUCCCC__AC___________________GGCGGUCGCAUUUGCAUAACAGCCGGUCG____ ..(((((..((((((..........))))))(((..(((.....)))(((((..(((((............................)))))..))))).........)))))))).... (-30.15 = -30.79 + 0.64)



| Location | 18,101,075 – 18,101,168 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -23.96 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18101075 93 + 22224390 UGGCAGGCCGAUGGCUGAUCUCUCGCCAUCUUGGUCGCCGUCACUGCAGCACUGCACCACCACCUCCAUCAGCACCGCCACCUUCUGCUCCUG ((((.(((.((((((.((((............))))))))))..((((....))))...............)).).))))............. ( -24.10) >DroSec_CAF1 9703 90 + 1 UGGCAGGCCGAUGGCUGAUCUCUCGCCAUCUUGGUCGCCGCCACUGCAGCACUGCACCACCUCCUCCA---CCACCGCCACCUUCUGCUGCUG ((((.((((((((((.((....))))))))..))))...))))..((((((.................---..............)))))).. ( -26.48) >DroSim_CAF1 9777 90 + 1 UGGCAGGCCGAUGGCUGAUCUCUCGCCAUCUUGGUCGCCGCCACUGCAGCACUGCACCACCACCUCCA---GCACCGCCACCUUCUGCUGCUG ((((.((((((((((.((....))))))))..))))...)))).((((....))))..........((---(((..((........))))))) ( -28.30) >DroEre_CAF1 7155 83 + 1 UGGCAGGCCGAUGGCUGAUCUCUCGCCAUCUUGGUCGCCGCCACUGCUGCACUGCAGCACC---GC-A---GCACCGCCACCA---ACUCCUG ((((.((((((((((.((....))))))))..))))((.((...((((((...))))))..---))-.---))...))))...---....... ( -33.40) >consensus UGGCAGGCCGAUGGCUGAUCUCUCGCCAUCUUGGUCGCCGCCACUGCAGCACUGCACCACCACCUCCA___GCACCGCCACCUUCUGCUCCUG ((((.((((((((((.((....))))))))..))))((.((....)).))..........................))))............. (-23.96 = -23.78 + -0.19)


| Location | 18,101,075 – 18,101,168 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -37.25 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18101075 93 - 22224390 CAGGAGCAGAAGGUGGCGGUGCUGAUGGAGGUGGUGGUGCAGUGCUGCAGUGACGGCGACCAAGAUGGCGAGAGAUCAGCCAUCGGCCUGCCA .............((((((.((((((((.(((.((.((.((.((...)).)))).)).)))....((((....).))).)))))))))))))) ( -37.10) >DroSec_CAF1 9703 90 - 1 CAGCAGCAGAAGGUGGCGGUGG---UGGAGGAGGUGGUGCAGUGCUGCAGUGGCGGCGACCAAGAUGGCGAGAGAUCAGCCAUCGGCCUGCCA ...........(((((((((.(---(.(..(..(..((.....))..)..)..).)).)))..((((((((....)).)))))).))).))). ( -33.90) >DroSim_CAF1 9777 90 - 1 CAGCAGCAGAAGGUGGCGGUGC---UGGAGGUGGUGGUGCAGUGCUGCAGUGGCGGCGACCAAGAUGGCGAGAGAUCAGCCAUCGGCCUGCCA ..((((((....(((.((.(((---.....))).)).)))..))))))..((((((.(.((..((((((((....)).))))))))))))))) ( -36.20) >DroEre_CAF1 7155 83 - 1 CAGGAGU---UGGUGGCGGUGC---U-GC---GGUGCUGCAGUGCAGCAGUGGCGGCGACCAAGAUGGCGAGAGAUCAGCCAUCGGCCUGCCA .......---((((((((((((---(-((---.(((((((...)))))).).))))).)))..((((((((....)).)))))).))).)))) ( -41.80) >consensus CAGCAGCAGAAGGUGGCGGUGC___UGGAGGUGGUGGUGCAGUGCUGCAGUGGCGGCGACCAAGAUGGCGAGAGAUCAGCCAUCGGCCUGCCA ...........(((((((((.............(.....)..((((((....)))))))))..((((((((....)).)))))).))).))). (-26.98 = -27.48 + 0.50)

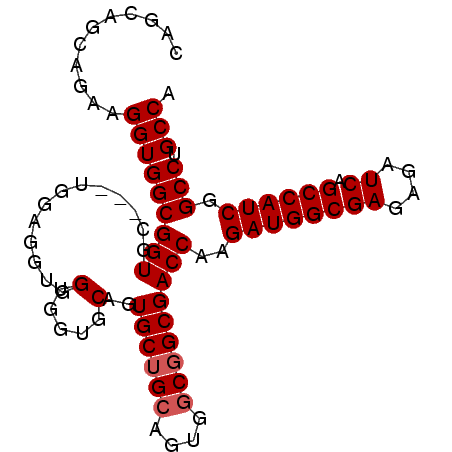
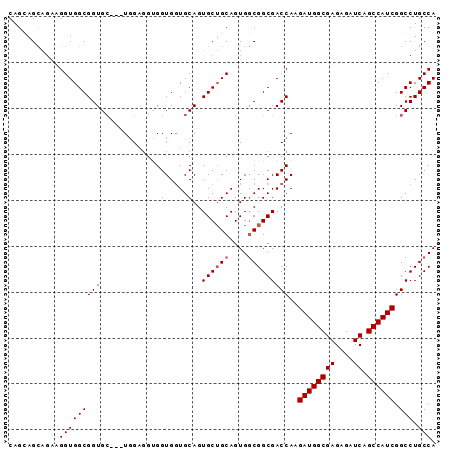
| Location | 18,101,168 – 18,101,267 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -33.94 |
| Energy contribution | -33.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18101168 99 - 22224390 UCUGCUUCGCUCUCGCUCGCUCUAAUCUGGGGAAUGCCAUCGCAGGAGCCACAGAACAAUGGCUCAGCGGAUCAGGAUCUCCGAAGGGAGGAGCAGGAG .........(((..((((.((((..((.(((((...((.((((..((((((........)))))).))))....)).)))))))..))))))))..))) ( -38.90) >DroSec_CAF1 9793 99 - 1 UCUGCUUCGCUCUCGCUCGCUCUAAUCUGGGGAAUGCCAUCGCAGGAGCCACAGAACAAUGGCUCAGCGGAUCAGGAUCUCCGAAGGGAGGAGCAGGAG .........(((..((((.((((..((.(((((...((.((((..((((((........)))))).))))....)).)))))))..))))))))..))) ( -38.90) >DroSim_CAF1 9867 99 - 1 UCUGCUUCGCUCUCGCUCGCUCUAAUCUGGGGAAUGCCAUCGCAGGAGCCACAGAACAAUGGCUCAGCGGAUCAGGAUCUCCGAAGGGAGGAGCAGGAG .........(((..((((.((((..((.(((((...((.((((..((((((........)))))).))))....)).)))))))..))))))))..))) ( -38.90) >DroYak_CAF1 8950 93 - 1 UCUGCUACGCUCUCGCUCGCUCUAAUCUGGGGAAUGCCAUCGCAGGAGCCACAGAACAAUGGCUCAG--GAU----AUCUUCAAAUGGAGGGGCAGGAG .........(((..((((.(((((...((((((.(((....))).((((((........))))))..--...----.))))))..)))))))))..))) ( -31.70) >consensus UCUGCUUCGCUCUCGCUCGCUCUAAUCUGGGGAAUGCCAUCGCAGGAGCCACAGAACAAUGGCUCAGCGGAUCAGGAUCUCCGAAGGGAGGAGCAGGAG .........(((..((((.((((....((((((.(((....))).((((((........))))))............))))))...))))))))..))) (-33.94 = -33.38 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:28 2006