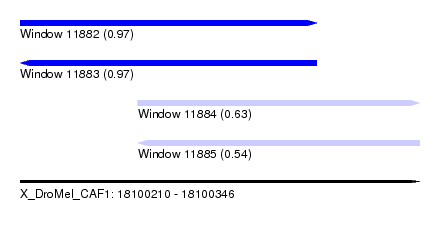
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,100,210 – 18,100,346 |
| Length | 136 |
| Max. P | 0.973322 |

| Location | 18,100,210 – 18,100,311 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.03 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -19.08 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18100210 101 + 22224390 UGUGUAUUUAAUUUAUGUGCUCGACGUGACACUUGCCAAGGGCCCCUGGCAUUGG--CGAUGGUGGUUCUUGGC--UGGU------AUGAGUAUGCAUAUUAUAGCAU---------AUA ((((((((((..((((((.....)))))).(((.(((((((((((((.((....)--).).)).))))))))))--.)))------.))))))))))...........---------... ( -34.60) >DroSec_CAF1 8867 101 + 1 UGUGUAUUUAAUUUAUGUGCUCGACGUGACACUUGCCAAGGGCCCCUGGCAUUGG--CGAUGGUGGUUCUUGGC--UGGU------AUGGGUAUGCAUAUUAUAGCAU---------AUA ((((((((((..((((((.....)))))).(((.(((((((((((((.((....)--).).)).))))))))))--.)))------.))))))))))...........---------... ( -33.70) >DroSim_CAF1 8923 101 + 1 UGUGUAUUUAAUUUAUGUGCUCGACGUGACACUUGCCAAGGGCCCCUGGCAUUGG--CGAUGGUGGUUCUUGGC--UGGU------AUGGGUAUGCAUAUUAUAGCAU---------AUA ((((((((((..((((((.....)))))).(((.(((((((((((((.((....)--).).)).))))))))))--.)))------.))))))))))...........---------... ( -33.70) >DroEre_CAF1 6361 76 + 1 UAUGUUACUAAUUUAUGUGCCUGGCGUGACACUUGCCAGGGGCUCCUGGCACUGG--CGAUGGUGGUUCCUGGA--UGGU---------------------------------------- ......((((......((.(((((((.......))))))).))(((.((((((..--....))))...)).)))--))))---------------------------------------- ( -23.80) >DroYak_CAF1 8007 116 + 1 UGUGUAUUUAAUUUAUGUGCUUGACGUGACACUUGCCAAGGGCCUCCGGCAUUGG--CGAUGGUGGUUCUGGGC--UGGUAAUGGUAUGGGUAUAUAUAUUAUAGCAUAUGGUAUAUAUA (((((((.....(((((((((.........(((.(((.((((((.(((((....)--)..))).)))))).)))--.))).((((((((......))))))))))))))))).))))))) ( -34.00) >DroAna_CAF1 11145 108 + 1 UGUGUAUUUAAUUUAUGUGGUUGACGUGACACUUGCCGGGGGCCUCCGGGGGUGGACCGACCG----ACUUGGCCCUGGC------AUUGGUCCAAAAA-GGCGGCAAGUGGCA-AGGUA ..(((.......((((((.....))))))(((((((((((((((..(((.((....))..)))----....))))))(((------....)))......-..))))))))))))-..... ( -39.70) >consensus UGUGUAUUUAAUUUAUGUGCUCGACGUGACACUUGCCAAGGGCCCCUGGCAUUGG__CGAUGGUGGUUCUUGGC__UGGU______AUGGGUAUGCAUAUUAUAGCAU_________AUA ((((((((((..((((((.....))))))((...((((((((((...(.(((((...))))).)))))))))))..)).........))))))))))....................... (-19.08 = -21.00 + 1.92)



| Location | 18,100,210 – 18,100,311 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.03 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18100210 101 - 22224390 UAU---------AUGCUAUAAUAUGCAUACUCAU------ACCA--GCCAAGAACCACCAUCG--CCAAUGCCAGGGGCCCUUGGCAAGUGUCACGUCGAGCACAUAAAUUAAAUACACA ..(---------((((........))))).....------....--((((((..((.((...(--(....))..))))..))))))..((((........))))................ ( -23.10) >DroSec_CAF1 8867 101 - 1 UAU---------AUGCUAUAAUAUGCAUACCCAU------ACCA--GCCAAGAACCACCAUCG--CCAAUGCCAGGGGCCCUUGGCAAGUGUCACGUCGAGCACAUAAAUUAAAUACACA ..(---------((((........))))).....------....--((((((..((.((...(--(....))..))))..))))))..((((........))))................ ( -23.10) >DroSim_CAF1 8923 101 - 1 UAU---------AUGCUAUAAUAUGCAUACCCAU------ACCA--GCCAAGAACCACCAUCG--CCAAUGCCAGGGGCCCUUGGCAAGUGUCACGUCGAGCACAUAAAUUAAAUACACA ..(---------((((........))))).....------....--((((((..((.((...(--(....))..))))..))))))..((((........))))................ ( -23.10) >DroEre_CAF1 6361 76 - 1 ----------------------------------------ACCA--UCCAGGAACCACCAUCG--CCAGUGCCAGGAGCCCCUGGCAAGUGUCACGCCAGGCACAUAAAUUAGUAACAUA ----------------------------------------.((.--....))...........--....(((((((....))))))).(((((......)))))................ ( -19.80) >DroYak_CAF1 8007 116 - 1 UAUAUAUACCAUAUGCUAUAAUAUAUAUACCCAUACCAUUACCA--GCCCAGAACCACCAUCG--CCAAUGCCGGAGGCCCUUGGCAAGUGUCACGUCAAGCACAUAAAUUAAAUACACA ((((((((..(((...)))..))))))))...............--(((.((..((.((...(--(....)).)).))..)).)))..((((........))))................ ( -16.30) >DroAna_CAF1 11145 108 - 1 UACCU-UGCCACUUGCCGCC-UUUUUGGACCAAU------GCCAGGGCCAAGU----CGGUCGGUCCACCCCCGGAGGCCCCCGGCAAGUGUCACGUCAACCACAUAAAUUAAAUACACA .....-((.(((((((((..-....(((......------.)))(((((...(----(((..((....)).)))).))))).))))))))).)).......................... ( -34.70) >consensus UAU_________AUGCUAUAAUAUGCAUACCCAU______ACCA__GCCAAGAACCACCAUCG__CCAAUGCCAGGGGCCCUUGGCAAGUGUCACGUCAAGCACAUAAAUUAAAUACACA .....................................................................(((((((....))))))).((((........))))................ (-12.15 = -12.40 + 0.25)


| Location | 18,100,250 – 18,100,346 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -27.23 |
| Energy contribution | -27.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18100250 96 + 22224390 GGCCCCUGGCAUUGGCGAUGGUGGUUCUUGGCUGGUAUGAGUAUGCAUAUUAUAGCAUAUAGCAUAUCGGCAUUGGCUGGUUAGUGUGGUAAGUGU .(((((((((...(.((((((....))...(((((((((.((((((........))))))..))))))))))))).)..))))).).)))...... ( -29.80) >DroSec_CAF1 8907 89 + 1 GGCCCCUGGCAUUGGCGAUGGUGGUUCUUGGCUGGUAUGGGUAUGCAUAUUAUAGCAUAUAGCAUAUCGGCAUUGGCUGGUAAGUGUGG------- (((((((.((....)).).)).))))....(((((((((.((((((........))))))..)))))))))....(((....)))....------- ( -27.80) >DroSim_CAF1 8963 89 + 1 GGCCCCUGGCAUUGGCGAUGGUGGUUCUUGGCUGGUAUGGGUAUGCAUAUUAUAGCAUAUAGCAUAUCGGCAUUGGCUGGUAAGUGUGG------- (((((((.((....)).).)).))))....(((((((((.((((((........))))))..)))))))))....(((....)))....------- ( -27.80) >consensus GGCCCCUGGCAUUGGCGAUGGUGGUUCUUGGCUGGUAUGGGUAUGCAUAUUAUAGCAUAUAGCAUAUCGGCAUUGGCUGGUAAGUGUGG_______ (((((((.((....)).).)).........(((((((((.((((((........))))))..)))))))))...)))).................. (-27.23 = -27.23 + -0.00)

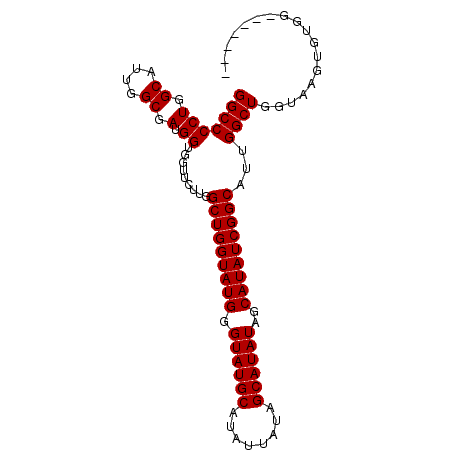

| Location | 18,100,250 – 18,100,346 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -14.70 |
| Consensus MFE | -14.70 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18100250 96 - 22224390 ACACUUACCACACUAACCAGCCAAUGCCGAUAUGCUAUAUGCUAUAAUAUGCAUACUCAUACCAGCCAAGAACCACCAUCGCCAAUGCCAGGGGCC ...................(((...((.(.((((...(((((........)))))..)))).).)).........((...((....))..))))). ( -14.70) >DroSec_CAF1 8907 89 - 1 -------CCACACUUACCAGCCAAUGCCGAUAUGCUAUAUGCUAUAAUAUGCAUACCCAUACCAGCCAAGAACCACCAUCGCCAAUGCCAGGGGCC -------............(((...((.(.((((...(((((........)))))..)))).).)).........((...((....))..))))). ( -14.70) >DroSim_CAF1 8963 89 - 1 -------CCACACUUACCAGCCAAUGCCGAUAUGCUAUAUGCUAUAAUAUGCAUACCCAUACCAGCCAAGAACCACCAUCGCCAAUGCCAGGGGCC -------............(((...((.(.((((...(((((........)))))..)))).).)).........((...((....))..))))). ( -14.70) >consensus _______CCACACUUACCAGCCAAUGCCGAUAUGCUAUAUGCUAUAAUAUGCAUACCCAUACCAGCCAAGAACCACCAUCGCCAAUGCCAGGGGCC ...................(((...((.(.((((...(((((........)))))..)))).).)).........((...((....))..))))). (-14.70 = -14.70 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:24 2006