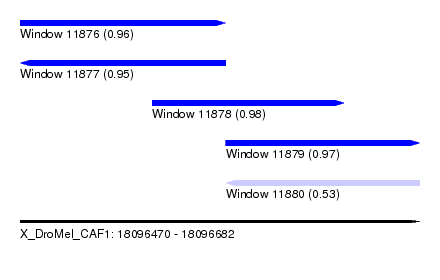
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,096,470 – 18,096,682 |
| Length | 212 |
| Max. P | 0.984090 |

| Location | 18,096,470 – 18,096,579 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -19.90 |
| Energy contribution | -22.77 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18096470 109 + 22224390 CAGCAACAAUAACAUGCGAUAGCUGCAACAACAACUGCUCAGAGCCAUAAUAAACAAACAAUUACGGUUUCAUUUUGUUGCAGUUGCAGCCCGGCAGUGGAAGUUGCUG (((((((....((.(((....((((((((..((((......(((((.((((.........)))).)))))......))))..))))))))...)))))....))))))) ( -35.80) >DroSec_CAF1 5083 109 + 1 CAGCAACAAUAACAUGCGAUAGCUGCAACAACAACUGCUCAGAGUCAUAAUAAACAAACAAUUACGGUUUCAUUUUGUUGCAGUUGCAGUCCGGCAGUGGAAGUUGCUG (((((((....((.(((....((((((((..((((......((..(.((((.........)))).)..))......))))..))))))))...)))))....))))))) ( -29.90) >DroSim_CAF1 4628 109 + 1 CAGCAACAAUAACAUGCGAUAGCUGCAACAACAACUGCUCAGAGCCAUAAUAAACAAACAAUUACGGUUUCAUUUUGUUGCAGUUGCAGUCCGGCAGUGGAAGUUGCUG (((((((....((.(((....((((((((..((((......(((((.((((.........)))).)))))......))))..))))))))...)))))....))))))) ( -34.20) >DroEre_CAF1 3278 89 + 1 CAACAACAGCAACAUGCGAUUGCCACAGCAACAACUGCACAGAGCCAUAAUAAACGAACAAUUACGCUUUCAUUUUGUUGGAGCU--------------------GCAG ...(((..((.....))..)))...((((..((((.....(((((..((((.........)))).)))))......))))..)))--------------------)... ( -15.40) >consensus CAGCAACAAUAACAUGCGAUAGCUGCAACAACAACUGCUCAGAGCCAUAAUAAACAAACAAUUACGGUUUCAUUUUGUUGCAGUUGCAGUCCGGCAGUGGAAGUUGCUG (((((((.......(((....((((((((..((((......(((((.((((.........)))).)))))......))))..))))))))...)))......))))))) (-19.90 = -22.77 + 2.87)



| Location | 18,096,470 – 18,096,579 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -21.02 |
| Energy contribution | -22.78 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18096470 109 - 22224390 CAGCAACUUCCACUGCCGGGCUGCAACUGCAACAAAAUGAAACCGUAAUUGUUUGUUUAUUAUGGCUCUGAGCAGUUGUUGUUGCAGCUAUCGCAUGUUAUUGUUGCUG (((((((....(((((..(((((((((.(((((.....((..(((((((.........))))))).))......))))).)))))))))...))).))....))))))) ( -40.20) >DroSec_CAF1 5083 109 - 1 CAGCAACUUCCACUGCCGGACUGCAACUGCAACAAAAUGAAACCGUAAUUGUUUGUUUAUUAUGACUCUGAGCAGUUGUUGUUGCAGCUAUCGCAUGUUAUUGUUGCUG (((((((....(((((.((.(((((((.(((((...(((....)))...((((((((......)))...)))))))))).)))))))))...))).))....))))))) ( -32.90) >DroSim_CAF1 4628 109 - 1 CAGCAACUUCCACUGCCGGACUGCAACUGCAACAAAAUGAAACCGUAAUUGUUUGUUUAUUAUGGCUCUGAGCAGUUGUUGUUGCAGCUAUCGCAUGUUAUUGUUGCUG (((((((....(((((.((.(((((((.(((((.....((..(((((((.........))))))).))......))))).)))))))))...))).))....))))))) ( -34.90) >DroEre_CAF1 3278 89 - 1 CUGC--------------------AGCUCCAACAAAAUGAAAGCGUAAUUGUUCGUUUAUUAUGGCUCUGUGCAGUUGUUGCUGUGGCAAUCGCAUGUUGCUGUUGUUG ..((--------------------(((..((((...(((((((((........))))).)))).((.....)).))))..)))))((((((.((.....)).)))))). ( -24.00) >consensus CAGCAACUUCCACUGCCGGACUGCAACUGCAACAAAAUGAAACCGUAAUUGUUUGUUUAUUAUGGCUCUGAGCAGUUGUUGUUGCAGCUAUCGCAUGUUAUUGUUGCUG (((((((.(((......)))(((((((..((((..........((((((.........))))))((.....)).))))..)))))))...............))))))) (-21.02 = -22.78 + 1.75)

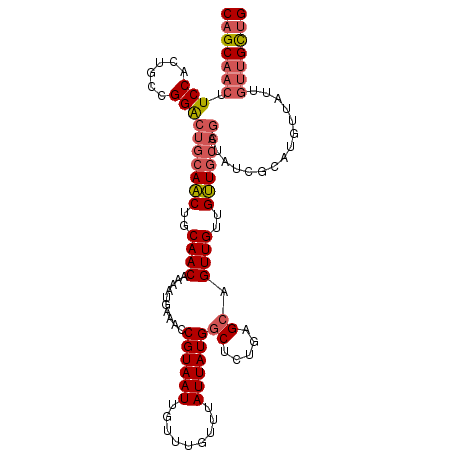

| Location | 18,096,540 – 18,096,642 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.14 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18096540 102 + 22224390 CAUUUUGUUGCA-GUUGCAGCCCGGCAGUGGAAGUUGCUGCCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAG-UGCCACU---UGGGCCUG ......(((((.-...)))))..(((((..(...)..))))).((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).. ( -44.70) >DroSec_CAF1 5153 102 + 1 CAUUUUGUUGCA-GUUGCAGUCCGGCAGUGGAAGUUGCUGCCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAG-UGCCACU---UGGGCCUG .......((((.-...))))...(((((..(...)..))))).((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).. ( -41.70) >DroSim_CAF1 4698 102 + 1 CAUUUUGUUGCA-GUUGCAGUCCGGCAGUGGAAGUUGCUGCCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAA-UGCCACU---UGGGCCUG .......((((.-...))))...(((((..(...)..))))).((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).. ( -41.70) >DroEre_CAF1 3348 82 + 1 CAUUUUGUUGGA-GCU--------------------GCAGCCUGGCCCUGGUACUGCCGCA--------GUUGCA-----GGUCUCCAUCUUGGCUAGCAG-UGCCACA---UGGGCCUG ......((((..-...--------------------.))))..((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).. ( -32.50) >DroYak_CAF1 4033 102 + 1 CAUUUUGUUGCA-GUUGCGGCCUGGCAGUGGAAGUUGCAACCUGGCCCUGGUACUGCAGCA--------GUUGCA-----UGUCUCCAUAUUGGCUGGCAG-UGCCACU---UGGGCCUG ......((((((-...((..((.......))..))))))))..((((((((((((((.((.--------...)).-----.(((........)))..))))-)))))..---.))))).. ( -39.40) >DroAna_CAF1 6357 112 + 1 CAUUUUGUUGCACGUUGCAA---GGCAAUGGU-GUUGU----UGGUGUUGGUGUUGCAGUGGCACUUUUGUCGCAAAAACAGUCUCCAUAUUGGCUGGCAAAUACUAUAUACUGUAUAUA ((((.(.((((.....))))---.).))))((-((...----(((((((((((((((.((((((....)))))).....(((((........))))))).))))))).)))))).)))). ( -32.20) >consensus CAUUUUGUUGCA_GUUGCAG_CCGGCAGUGGAAGUUGCUGCCUGGCCCUGGUAUUGCAGCA________GUUGCA_____AGUCUCCAUAUUGGCUAGCAG_UGCCACU___UGGGCCUG ..(((..((((.............))))..)))..........((((((((((((((.(((..........)))......((((........)))).)))).)))))......))))).. (-23.63 = -23.14 + -0.49)



| Location | 18,096,579 – 18,096,682 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -42.39 |
| Consensus MFE | -32.14 |
| Energy contribution | -30.62 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18096579 103 + 22224390 CCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAG-UGCCACU---UGGGCCUGCCCCGCCCCCGAAUGCCCCACCAGGGGGCACUGCUCCGCA ...((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).((...((((((.............))))))...))...... ( -43.62) >DroSec_CAF1 5192 103 + 1 CCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAG-UGCCACU---UGGGCCUGCCCCGCCCCCAAAUGCCCCACCAGGGGGCACCGCUCUGCA ...((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).((...((((((.............))))))...))...... ( -42.92) >DroSim_CAF1 4737 102 + 1 CCUGGCCCUGGUAUUGCAGCA--------GUUGCA-----AGUCUCCAUAUUGGCUAGCAA-UGCCACU---UGGGCCUGCCCCGCCCCCAAAUGCCCCACCAGG-GGCACCACUCCGCA ...((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.)))))......((.......((((((....))-)))).......)). ( -42.14) >DroEre_CAF1 3367 103 + 1 CCUGGCCCUGGUACUGCCGCA--------GUUGCA-----GGUCUCCAUCUUGGCUAGCAG-UGCCACA---UGGGCCUGCCCCGCCCCCACAUUCCCCACCAGGGGGCACAGCUCCGGA ((.((((((((((((((.((.--------...)).-----((((........)))).))))-)))))..---.))))).((...((((((.............))))))...))...)). ( -45.72) >DroYak_CAF1 4072 96 + 1 CCUGGCCCUGGUACUGCAGCA--------GUUGCA-----UGUCUCCAUAUUGGCUGGCAG-UGCCACU---UGGGCCUGCCCCGCCCCCAAAUGCCCCACCAGGGGGCACUG------- ...((((((((((((((.((.--------...)).-----.(((........)))..))))-)))))..---.)))))......((((((.............))))))....------- ( -43.02) >DroAna_CAF1 6391 118 + 1 --UGGUGUUGGUGUUGCAGUGGCACUUUUGUCGCAAAAACAGUCUCCAUAUUGGCUGGCAAAUACUAUAUACUGUAUAUACCCCUGCCUCCUAAGCCCCACCAGGGGGCACUGCCACAGA --((((...((((((((.((((((....)))))).....(((((........)))))))))((((........)))).))))..((((((((..........))))))))..)))).... ( -36.90) >consensus CCUGGCCCUGGUAUUGCAGCA________GUUGCA_____AGUCUCCAUAUUGGCUAGCAG_UGCCACU___UGGGCCUGCCCCGCCCCCAAAUGCCCCACCAGGGGGCACUGCUCCGCA ...((((((((((((((.(((..........)))......((((........)))).)))).)))))......)))))......((((((.............))))))........... (-32.14 = -30.62 + -1.52)

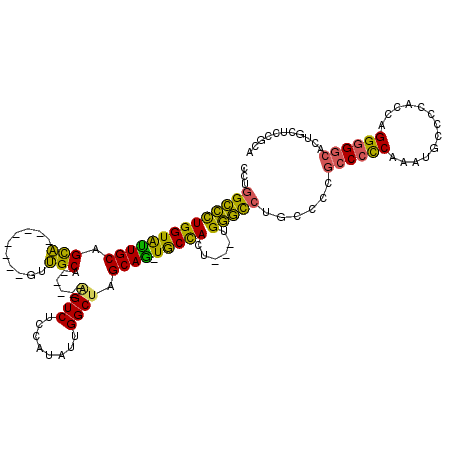
| Location | 18,096,579 – 18,096,682 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -43.12 |
| Consensus MFE | -28.57 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18096579 103 - 22224390 UGCGGAGCAGUGCCCCCUGGUGGGGCAUUCGGGGGCGGGGCAGGCCCA---AGUGGCA-CUGCUAGCCAAUAUGGAGACU-----UGCAAC--------UGCUGCAAUACCAGGGCCAGG ......((..(((((((..(((...)))..)))))))..)).(((((.---..((((.-......))))...(((....(-----((((..--------...)))))..))))))))... ( -43.60) >DroSec_CAF1 5192 103 - 1 UGCAGAGCGGUGCCCCCUGGUGGGGCAUUUGGGGGCGGGGCAGGCCCA---AGUGGCA-CUGCUAGCCAAUAUGGAGACU-----UGCAAC--------UGCUGCAAUACCAGGGCCAGG ......((..(((((((..(((...)))..)))))))..)).(((((.---..((((.-......))))...(((....(-----((((..--------...)))))..))))))))... ( -42.90) >DroSim_CAF1 4737 102 - 1 UGCGGAGUGGUGCC-CCUGGUGGGGCAUUUGGGGGCGGGGCAGGCCCA---AGUGGCA-UUGCUAGCCAAUAUGGAGACU-----UGCAAC--------UGCUGCAAUACCAGGGCCAGG ...((..(((((((-((....))))).....(.(((((.(((((((((---..((((.-......))))...))).).))-----)))..)--------)))).)...))))...))... ( -42.70) >DroEre_CAF1 3367 103 - 1 UCCGGAGCUGUGCCCCCUGGUGGGGAAUGUGGGGGCGGGGCAGGCCCA---UGUGGCA-CUGCUAGCCAAGAUGGAGACC-----UGCAAC--------UGCGGCAGUACCAGGGCCAGG .((...(((.(((((((..((.....))..))))))).))).(((((.---..(((.(-(((((..((.....)).....-----.((...--------.)))))))).)))))))).)) ( -47.90) >DroYak_CAF1 4072 96 - 1 -------CAGUGCCCCCUGGUGGGGCAUUUGGGGGCGGGGCAGGCCCA---AGUGGCA-CUGCCAGCCAAUAUGGAGACA-----UGCAAC--------UGCUGCAGUACCAGGGCCAGG -------...(((((((..(((...)))..))))))).....(((((.---..(((.(-((((.(((...((((....))-----))....--------.)))))))).))))))))... ( -45.50) >DroAna_CAF1 6391 118 - 1 UCUGUGGCAGUGCCCCCUGGUGGGGCUUAGGAGGCAGGGGUAUAUACAGUAUAUAGUAUUUGCCAGCCAAUAUGGAGACUGUUUUUGCGACAAAAGUGCCACUGCAACACCAACACCA-- ..(((.((((((((((.....)))))......((((..((((.((((........)))).)))).((.((((.(....)))))...))........))))))))).))).........-- ( -36.10) >consensus UGCGGAGCAGUGCCCCCUGGUGGGGCAUUUGGGGGCGGGGCAGGCCCA___AGUGGCA_CUGCUAGCCAAUAUGGAGACU_____UGCAAC________UGCUGCAAUACCAGGGCCAGG ..............((((((((..((.......(((..((((((((........)))..))))).))).....(....)........................))..))))))))..... (-28.57 = -29.05 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:20 2006