
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,092,704 – 18,092,861 |
| Length | 157 |
| Max. P | 0.954786 |

| Location | 18,092,704 – 18,092,821 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -29.14 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18092704 117 + 22224390 GUGGGAUAUAAAUUCAGUCGUUUCUGGGCUAUUUCCACAGAGACAUC---UCAAAGAGAACCCUACCAAGCCUUGCACUGAUCUAUCUAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUA ..(((..............(((((((((......)).))))))).((---((...)))).)))((((((((((.(((..(((((((...........)))))))..))).)))))))))) ( -45.20) >DroSec_CAF1 1421 113 + 1 GUGGUAUAUAAAUUCAGUCGUUGCUGGGCUAUUUCCACAGAGACAUC---UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC----UAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUA ((((...(((..((((((....)))))).)))..))))((((...((---((...)))).))))(((((((((.(((..((((----(((.......)))))))..))).))))))))). ( -41.80) >DroSim_CAF1 1414 113 + 1 GUGGGAUAUAAAUUCAGUCGUUGCUGGGCUAUUUCCACAGAGACAUC---UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC----UAUACAAAUCGUAGAUCUCUGCCAGGCUUGGUA ((((((......((((((....))))))....))))))((((...((---((...)))).))))(((((((((.(((..((((----(((.......)))))))..))).))))))))). ( -43.40) >DroYak_CAF1 147 116 + 1 GUGGGAUAUAAAUUCAGUCGUUGCUGGGCGAUUUCCACAGAGACAUCCUCCUAAAGAUAACUCUCUCAAGCCUUGCACAGAUC----UAUACAUAUCAUAGAUCUCUGCCAGACUUGGUA ((((((......((((((....))))))....))))))(((((.(((........)))...)))))((((.((.(((.(((((----(((.......)))))))).))).)).))))... ( -32.50) >consensus GUGGGAUAUAAAUUCAGUCGUUGCUGGGCUAUUUCCACAGAGACAUC___UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC____UAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUA ((((((......(((((......)))))....))))))((((...((........))...))))(((((((((.(((..((((.................))))..))).))))))))). (-29.14 = -29.96 + 0.81)



| Location | 18,092,704 – 18,092,821 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18092704 117 - 22224390 UACCAAGCCUGGCAGAGAUCUACGAUAUGUAUAGAUAGAUCAGUGCAAGGCUUGGUAGGGUUCUCUUUGA---GAUGUCUCUGUGGAAAUAGCCCAGAAACGACUGAAUUUAUAUCCCAC ((((((((((.(((..(((((((..........).))))))..))).))))))))))(((......((.(---(.(((.((((.((......)))))).))).)).)).......))).. ( -42.92) >DroSec_CAF1 1421 113 - 1 UACCAAGCCUGGCAGAGAUCUACGAUAUGUAUA----GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA---GAUGUCUCUGUGGAAAUAGCCCAGCAACGACUGAAUUUAUAUACCAC ((((((((((.(((..((((((.........))----))))..))).))))))))))(((.((((...))---))...))).((((..((((..(((......)))...))))...)))) ( -37.70) >DroSim_CAF1 1414 113 - 1 UACCAAGCCUGGCAGAGAUCUACGAUUUGUAUA----GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA---GAUGUCUCUGUGGAAAUAGCCCAGCAACGACUGAAUUUAUAUCCCAC ((((((((((.(((..((((((.........))----))))..))).))))))))))(((.((((...))---))...)))((.(((.((((..(((......)))...)))).))))). ( -38.80) >DroYak_CAF1 147 116 - 1 UACCAAGUCUGGCAGAGAUCUAUGAUAUGUAUA----GAUCUGUGCAAGGCUUGAGAGAGUUAUCUUUAGGAGGAUGUCUCUGUGGAAAUCGCCCAGCAACGACUGAAUUUAUAUCCCAC ...(((((((.(((.(((((((((.....))))----))))).))).)))))))..((((.((((((....)))))).))))((((........(((......)))..........)))) ( -37.17) >consensus UACCAAGCCUGGCAGAGAUCUACGAUAUGUAUA____GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA___GAUGUCUCUGUGGAAAUAGCCCAGCAACGACUGAAUUUAUAUCCCAC ((((((((((.(((..((((.................))))..))).))))))))))((((((((........))(((..(((.((......)))))..)))...))))))......... (-27.70 = -28.08 + 0.38)

| Location | 18,092,744 – 18,092,861 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.96 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -5.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18092744 117 + 22224390 AGACAUC---UCAAAGAGAACCCUACCAAGCCUUGCACUGAUCUAUCUAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUAUCAUGCCUAACUACUUUGACUG .((.(((---(...(((((((.(((((((((((.(((..(((((((...........)))))))..))).))))))))))).)).)))))...)))).)).................... ( -43.10) >DroSec_CAF1 1461 113 + 1 AGACAUC---UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC----UAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUCUCAUGCAUAACUACUUUGGCUG .((.(((---(...(((((((.(((((((((((.(((..((((----(((.......)))))))..))).))))))))))).)).)))))...)))).)).................... ( -43.90) >DroSim_CAF1 1454 113 + 1 AGACAUC---UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC----UAUACAAAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUCUCAUGCCUAACUACUUUGGCUG .((.(((---(...(((((((.(((((((((((.(((..((((----(((.......)))))))..))).))))))))))).)).)))))...)))).))..(((..........))).. ( -47.30) >DroYak_CAF1 187 116 + 1 AGACAUCCUCCUAAAGAUAACUCUCUCAAGCCUUGCACAGAUC----UAUACAUAUCAUAGAUCUCUGCCAGACUUGGUAGCGCAUCUCCUCGGCAUCUCAUUCCUAACCACUUUGGCUG ..........((((((......((..((((.((.(((.(((((----(((.......)))))))).))).)).))))..)).((.........))................))))))... ( -24.00) >consensus AGACAUC___UCAAAGAGAACUCUACCAAGCCUUGCACCGAUC____UAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUCUCAUGCCUAACUACUUUGGCUG ..........((((((...((.(((((((((((.(((..((((.................))))..))).))))))))))).))((((.....))))..............))))))... (-27.70 = -27.96 + 0.25)

| Location | 18,092,744 – 18,092,861 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -32.38 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.77 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18092744 117 - 22224390 CAGUCAAAGUAGUUAGGCAUGAUAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGUAUAGAUAGAUCAGUGCAAGGCUUGGUAGGGUUCUCUUUGA---GAUGUCU ..(((..........)))..(((((((...(((((.((.(((((((((((.(((..(((((((..........).))))))..))).))))))))))).)))))))..).---)))))). ( -45.50) >DroSec_CAF1 1461 113 - 1 CAGCCAAAGUAGUUAUGCAUGAGAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGUAUA----GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA---GAUGUCU ........(((....)))..((.((((...(((((.((.(((((((((((.(((..((((((.........))----))))..))).))))))))))).)))))))..).---))).)). ( -44.10) >DroSim_CAF1 1454 113 - 1 CAGCCAAAGUAGUUAGGCAUGAGAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUUUGUAUA----GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA---GAUGUCU ..(((..........)))..((.((((...(((((.((.(((((((((((.(((..((((((.........))----))))..))).))))))))))).)))))))..).---))).)). ( -46.80) >DroYak_CAF1 187 116 - 1 CAGCCAAAGUGGUUAGGAAUGAGAUGCCGAGGAGAUGCGCUACCAAGUCUGGCAGAGAUCUAUGAUAUGUAUA----GAUCUGUGCAAGGCUUGAGAGAGUUAUCUUUAGGAGGAUGUCU .((((.....)))).......(((((((..((((((((.((..(((((((.(((.(((((((((.....))))----))))).))).)))))))..)).).)))))))....)).))))) ( -40.70) >consensus CAGCCAAAGUAGUUAGGCAUGAGAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGUAUA____GAUCGGUGCAAGGCUUGGUAGAGUUCUCUUUGA___GAUGUCU ..............((((((....((....(((((.((.(((((((((((.(((..((((.................))))..))).))))))))))).)))))))..))....)))))) (-32.38 = -33.13 + 0.75)

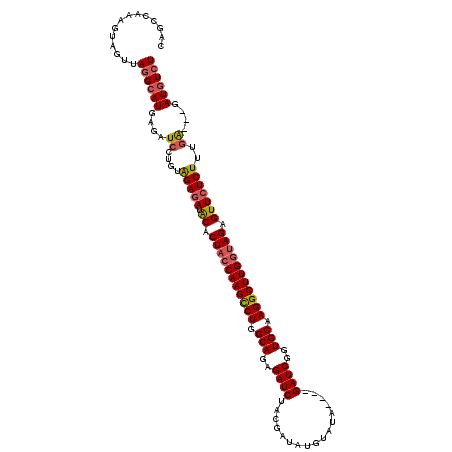

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:14 2006