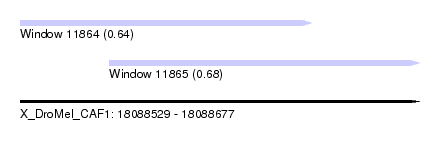
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,088,529 – 18,088,677 |
| Length | 148 |
| Max. P | 0.676573 |

| Location | 18,088,529 – 18,088,637 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18088529 108 + 22224390 UCUUAUCUUAUCGCCU-------UACUUUUCUCGCUGUGCAGCGGACACAAAUGCGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCGCUGC-----UCGUCUUAUCGC ................-------..........(.((.(((((((((((....(((..(((((((((....))))).)))))))...))))).....))))))-----.)).)....... ( -29.00) >DroSec_CAF1 10899 105 + 1 UCUUAUCUUAUCGCCU-------UUAUUUUCUCGCUGUGCAGCGAACACAAAUACGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---C-----CCGUCUUAUCGC ............((..-------.(((((..(((((....)))))....))))).)).(((((((((....))))).))))((..(((((....)))))---.-----.))......... ( -24.00) >DroSim_CAF1 10857 105 + 1 UCUUAUCUUAUCGCCU-------UUAUUUUCUCGCUGUGCAGCGAACACAAAUACGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---C-----CCGUCUUAUCGC ............((..-------.(((((..(((((....)))))....))))).)).(((((((((....))))).))))((..(((((....)))))---.-----.))......... ( -24.00) >DroEre_CAF1 8247 112 + 1 UCCUAUCUCAUCGCGAUAGCCGCUCCUUUUCUCGCGGUGCAGCGCGCUUCAAUGCGCCACCUGUUGCCAAUGCAACAAGGUCGUUCGGUGUUAUCACUG---C-----CCCUCUUAUCGC ............(((((((((((..........)))))((((.((((......))))((((((((((....)))))).((....))))))......)))---)-----......)))))) ( -36.00) >DroYak_CAF1 9856 108 + 1 UCUUAUCUUAUCGCGA---------CUUUUCUCGCUGUGCAGCGGCCUUAAAUGUGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---GUCGAUCGGUCUUAUCGC ............((((---------........(((....)))((((......).)))(((((((((....)))))).(((((.((((((....)))))---).))))))))....)))) ( -38.40) >consensus UCUUAUCUUAUCGCCU_______UUCUUUUCUCGCUGUGCAGCGAACACAAAUGCGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG___C_____CCGUCUUAUCGC ............((.................(((((....)))))..........((.(((((((((....))))).)))).)).(((((....)))))...................)) (-20.46 = -20.38 + -0.08)



| Location | 18,088,562 – 18,088,677 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -28.82 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
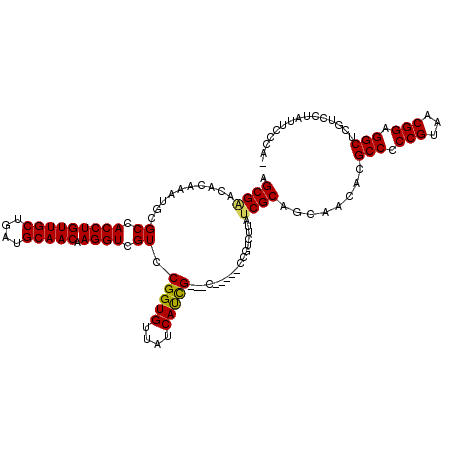
>X_DroMel_CAF1 18088562 115 + 22224390 AGCGGACACAAAUGCGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCGCUGC-----UCGUCUUAUCGCAGCAACACGCCCCCGUAACGGAGGCUCGUCCUAUUCCCAU ...((((......((((((((((((((....))))).)))).....))))).......(((((-----..........))))).....(((.(((...))).)))..))))......... ( -38.10) >DroSec_CAF1 10932 110 + 1 AGCGAACACAAAUACGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---C-----CCGUCUUAUCGCAGCAACACGCCCCCGUAACGGAGGCUCGUCCUAUUCCC-- .((((.............(((((((((....))))).))))((..(((((....)))))---.-----.)).....))))........(((.(((...))).))).............-- ( -31.20) >DroSim_CAF1 10890 110 + 1 AGCGAACACAAAUACGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---C-----CCGUCUUAUCGCAGCAACACGCCCCCGUAACGGAGGCUCGUCCUAUUCCC-- .((((.............(((((((((....))))).))))((..(((((....)))))---.-----.)).....))))........(((.(((...))).))).............-- ( -31.20) >DroEre_CAF1 8287 111 + 1 AGCGCGCUUCAAUGCGCCACCUGUUGCCAAUGCAACAAGGUCGUUCGGUGUUAUCACUG---C-----CCCUCUUAUCGCAGCA-CACGCCCCCGUAACGGAGGCUCGUCCUACUCCCAC .((((((......)))).(((((((((....))))).)))).(..(((((....)))))---.-----.)........))((.(-(..(((.(((...))).)))..)).))........ ( -32.00) >DroYak_CAF1 9887 117 + 1 AGCGGCCUUAAAUGUGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG---GUCGAUCGGUCUUAUCGCAGCAACACGCCCCCGUAACGGAGGCUCGUCCUAUUCCCAU .((((((......).)))(((((((((....)))))).(((((.((((((....)))))---).))))))))......))........(((.(((...))).)))............... ( -42.10) >consensus AGCGAACACAAAUGCGCCACCUGUUGCUGAUGCAACAAGGUCGUCCGGUGUUAUCAUCG___C_____CCGUCUUAUCGCAGCAACACGCCCCCGUAACGGAGGCUCGUCCUAUUCCCA_ .((((..........((.(((((((((....))))).)))).)).(((((....))))).................))))........(((.(((...))).)))............... (-28.82 = -28.54 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:07 2006