
| Sequence ID | X_DroMel_CAF1 |
|---|---|
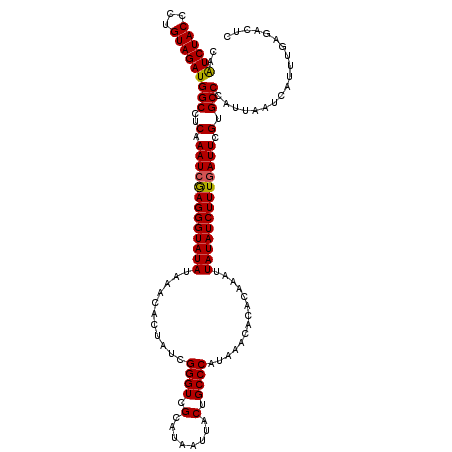
| Location | 18,084,918 – 18,085,036 |
| Length | 118 |
| Max. P | 0.999172 |

| Location | 18,084,918 – 18,085,036 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.29 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18084918 118 + 22224390 CAAUCUACCCUGUAGAUGGCCUCAAAUCGAGGGUAUAUAAACACUAUCGGGUCGCCUAAUUACUGCCCAUAAACACACAAAUUAUAUCUUUGAUUUGUGCCCAUUAAUCAUUUGAUAC-- ..((((((...))))))(((..(((((((((((((((...........((((.(........).))))..............))))))))))))))).))).(((((....)))))..-- ( -28.41) >DroSec_CAF1 7356 120 + 1 CAGUCUACCCUGUAGAUGGCCUCAAAUAGAGGGUAUAUAAACACUAUCGGGUCGCAUAAUUACUGCCCAUAAACACACAAAUUAUAUCUUUGAUUCGUGCCCAUUAAUCAUUUGAGACUC ..((((((...))))))((.(((((((.((((((((............((((.(........).)))).........((((.......))))....)))))).....))))))))).)). ( -24.20) >DroSim_CAF1 7132 120 + 1 CAGUCUACCCUGUAGAUGGCCUCAAAUCGAGGGUAUAUAAACACUAUCGGGUCGCAUAAUUACUGCCCAUAAACACACAAAUUAUAUCUUUGAUUCGUGCCCAUUAAUCAUUUGAGACUC .(((((....((..(((((.(.(.(((((((((((((...........((((.(........).))))..............))))))))))))).).).)))))...))....))))). ( -27.41) >DroEre_CAF1 4990 119 + 1 CAAUCUACCCUGUAGAUGGCCUCAAAUCAAGGGUAUAUA-ACACUAUCGGGUCGCCUAAUUACUGCCCAUAAGCACACAAAUUAUAUCUUUGAUUCGCGCCCAUUAAUCAUUUGAGACCU ..((((((...))))))((.(((((((....((((....-....))))(((.(((........(((......)))..((((.......))))....)))))).......))))))).)). ( -25.00) >DroYak_CAF1 5975 120 + 1 CCAUCUACCCUGUAGAUGGCCUCAAAUCAAGGGUAUAUAAACACUAUCGGGUCGCAUAAUCACUGCCCAUAAACACACAAAUUAUAUCUUUGAUUUGUGCCCAUUAAUCAUUUGAGACAU ((((((((...)))))))).(((((((...(((...............((((.(........).)))).......(((((((((......)))))))))))).......))))))).... ( -31.40) >consensus CAAUCUACCCUGUAGAUGGCCUCAAAUCGAGGGUAUAUAAACACUAUCGGGUCGCAUAAUUACUGCCCAUAAACACACAAAUUAUAUCUUUGAUUCGUGCCCAUUAAUCAUUUGAGACUC ..((((((...))))))(((..(.(((((((((((((...........((((.(........).))))..............))))))))))))).).)))................... (-23.57 = -23.29 + -0.28)

| Location | 18,084,918 – 18,085,036 |
|---|---|
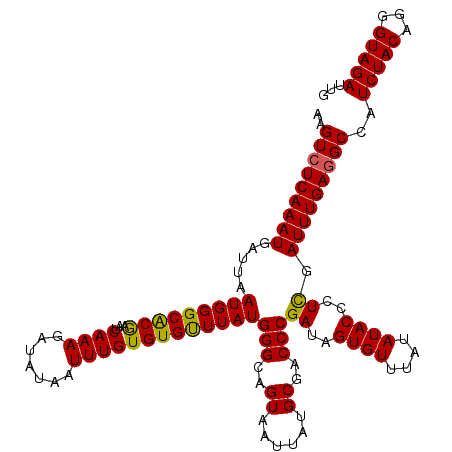
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -28.06 |
| Energy contribution | -27.46 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18084918 118 - 22224390 --GUAUCAAAUGAUUAAUGGGCACAAAUCAAAGAUAUAAUUUGUGUGUUUAUGGGCAGUAAUUAGGCGACCCGAUAGUGUUUAUAUACCCUCGAUUUGAGGCCAUCUACAGGGUAGAUUG --.((((...((.(..((((.(.((((((..(((((((.....)))))))..(((..((......))..)))((..((((....))))..)))))))).).))))..))).))))..... ( -26.00) >DroSec_CAF1 7356 120 - 1 GAGUCUCAAAUGAUUAAUGGGCACGAAUCAAAGAUAUAAUUUGUGUGUUUAUGGGCAGUAAUUAUGCGACCCGAUAGUGUUUAUAUACCCUCUAUUUGAGGCCAUCUACAGGGUAGACUG .((((...........((((((((..(((..(((((((.....)))))))..(((..(((....)))..)))))).)))))))).((((((.((..((....))..)).)))))))))). ( -31.40) >DroSim_CAF1 7132 120 - 1 GAGUCUCAAAUGAUUAAUGGGCACGAAUCAAAGAUAUAAUUUGUGUGUUUAUGGGCAGUAAUUAUGCGACCCGAUAGUGUUUAUAUACCCUCGAUUUGAGGCCAUCUACAGGGUAGACUG (.(((((((((((...((((((((..(((..(((((((.....)))))))..(((..(((....)))..)))))).))))))))......)).)))))))))).(((((...)))))... ( -30.50) >DroEre_CAF1 4990 119 - 1 AGGUCUCAAAUGAUUAAUGGGCGCGAAUCAAAGAUAUAAUUUGUGUGCUUAUGGGCAGUAAUUAGGCGACCCGAUAGUGU-UAUAUACCCUUGAUUUGAGGCCAUCUACAGGGUAGAUUG .((((((((((.(...(((((((((...((((.......)))))))))))))(((..((......))..)))........-..........).))))))))))((((((...)))))).. ( -33.80) >DroYak_CAF1 5975 120 - 1 AUGUCUCAAAUGAUUAAUGGGCACAAAUCAAAGAUAUAAUUUGUGUGUUUAUGGGCAGUGAUUAUGCGACCCGAUAGUGUUUAUAUACCCUUGAUUUGAGGCCAUCUACAGGGUAGAUGG ..(((((((((.(...((((((((..(((..(((((((.....)))))))..(((..((......))..)))))).)))))))).......).)))))))))(((((((...))))))). ( -32.60) >consensus AAGUCUCAAAUGAUUAAUGGGCACGAAUCAAAGAUAUAAUUUGUGUGUUUAUGGGCAGUAAUUAUGCGACCCGAUAGUGUUUAUAUACCCUCGAUUUGAGGCCAUCUACAGGGUAGAUUG ..(((((((((.....(((((((((...((((.......)))))))))))))(((..((......))..)))((..((((....))))..)).)))))))))..(((((...)))))... (-28.06 = -27.46 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:57 2006