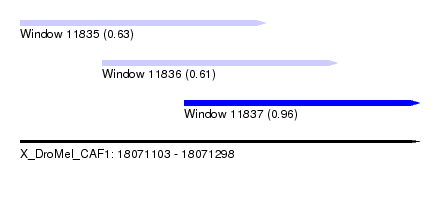
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,071,103 – 18,071,298 |
| Length | 195 |
| Max. P | 0.963524 |

| Location | 18,071,103 – 18,071,223 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -47.47 |
| Consensus MFE | -42.79 |
| Energy contribution | -42.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18071103 120 + 22224390 GGAUGAAGGGUGGCGUUGGGUGUCUGCGUCUGUUCUGCGCCCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGCCCUGUCGCCUCCAGAAUGCAUCCAACGCACCGCUAAUCCA ((((..((((((.((((((((((..(((.......)))((....(((((...)))))....)).......((((((((((......).))))))))))))))))))))))).)).)))). ( -48.00) >DroSec_CAF1 8777 120 + 1 GGAUGAAGGGUGGCGUUGGGUGUCUGCGUCUGUGCUGCACCCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUGCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCA ((((..((((((.((.(((((((...((...(((((........(((((...)))))...)))))...))(((((((((.((....)))))))))))))))))).)))))).)).)))). ( -49.10) >DroSim_CAF1 13366 120 + 1 GGAUGAAGGGUGGCGUUGGGUGUCUGCGUCUGUGCUGCACCCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUCCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCA ((((..((((((.((.(((((((...((...(((((........(((((...)))))...)))))...))(((((((((.(.....).)))))))))))))))).)))))).)).)))). ( -45.30) >consensus GGAUGAAGGGUGGCGUUGGGUGUCUGCGUCUGUGCUGCACCCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUCCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCA ((((..((((((.((.(((((((...((...(((((........(((((...)))))...)))))...))((((((((((......).)))))))))))))))).)))))).)).)))). (-42.79 = -42.90 + 0.11)

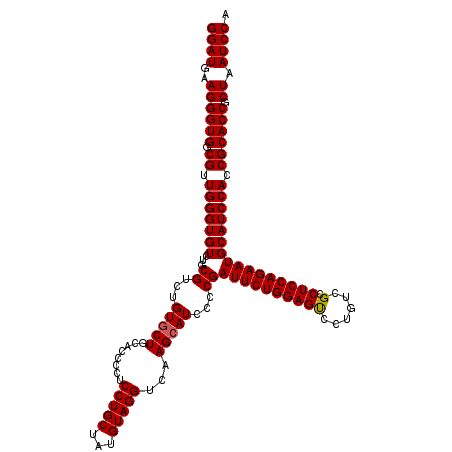
| Location | 18,071,143 – 18,071,258 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -22.65 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18071143 115 + 22224390 CCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGCCCUGUCGCCUCCAGAAUGCAUCCAACGCACCGCUAAUCCACAUGCCCACCCUCCGUGCAACCCGUC-----CACACGCCC .......((.((((.((...(((.......((((((((((......).)))))))))((.......))...)))...))))))))........((((.........-----..))))... ( -25.10) >DroSec_CAF1 8817 118 + 1 CCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUGCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA--CCGCCCAAAACACACUCCC .......((.((((.((...(((.......(((((((((.((....)))))))))))((.......))...)))...))))))))..............--................... ( -26.80) >DroSim_CAF1 13406 118 + 1 CCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUCCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA--CCGCCCAAAACACACUCCC .......((.((((.((...(((.......(((((((((.(.....).)))))))))((.......))...)))...))))))))..............--................... ( -23.00) >consensus CCCUCCUGCUAUGUAGGUCAAGCAUCCCCGAUUCUGGAGUCCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA__CCGCCCAAAACACACUCCC .......((.((((.((...(((.......((((((((((......).)))))))))((.......))...)))...))))))))................................... (-22.65 = -22.43 + -0.22)

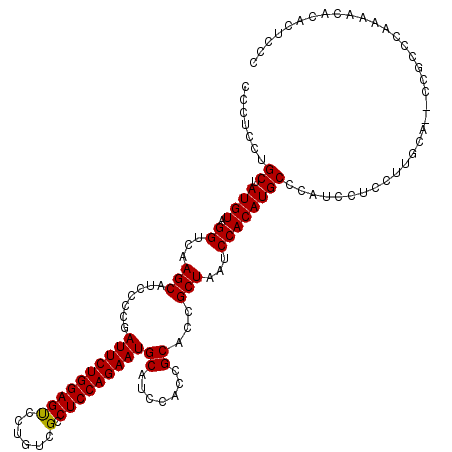
| Location | 18,071,183 – 18,071,298 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18071183 115 + 22224390 CCUGUCGCCUCCAGAAUGCAUCCAACGCACCGCUAAUCCACAUGCCCACCCUCCGUGCAACCCGUC-----CACACGCCCCGCGAAGGAUGCUCCACCUCAGCCCCGCCUGGUCGCGACA ...(((((..((((...((((((..(((...((..........))........((((.........-----..))))....)))..)))))).........((...))))))..))))). ( -33.30) >DroSec_CAF1 8857 118 + 1 GCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA--CCGCCCAAAACACACUCCCCGCGAAGGAUGCUCCACCCCACACCCGCCAGGUCGCGACA ...(((((..((.....((((((..(((...((..........))...........((.--..))................)))..)))))).........(....)...))..))))). ( -23.70) >DroSim_CAF1 13446 118 + 1 CCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA--CCGCCCAAAACACACUCCCCGCGAAGGAUGCUCCACCUCACACCCGCCUGGUCGCGACA ...(((((..((((...((((((..(((...((..........))...........((.--..))................)))..)))))).........(....).))))..))))). ( -27.40) >consensus CCUGUCGCCUCCAGAAUGCAUCCACCGCACCGCUAAUCCACAUGCCCAUCCUCCUUGCA__CCGCCCAAAACACACUCCCCGCGAAGGAUGCUCCACCUCACACCCGCCUGGUCGCGACA ...(((((..((((...((((((..(((...((..........))...........((.....))................)))..)))))).......(......).))))..))))). (-26.92 = -27.03 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:42 2006