
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,069,571 – 18,069,851 |
| Length | 280 |
| Max. P | 0.818109 |

| Location | 18,069,571 – 18,069,691 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -36.63 |
| Energy contribution | -36.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069571 120 + 22224390 UGGCAGCAAAUCAAUGGAUGGACUUAAAACCACCUGAAUUAGGUGCCUAAGGCUCCCAGUGCCACCCUGCUGUGUCCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGUAGGACGACUU .(((((((.(((....)))((((.......((((((...)))))).....(((.......)))..........))))))).))))(((((((((((((....))))))..)))))))... ( -39.20) >DroSec_CAF1 7311 119 + 1 UGGCAGCAAAUCAAUGGAAGGACUUAAAACCACCUGAAUUAGGUGCCUAAGGC-CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUU ..((((((.....((((.(((.........((((((...)))))).....(((-......)))..))).)))).)))))).....(((((((((((((....))))))..)))))))... ( -39.30) >DroSim_CAF1 11873 119 + 1 UGGCAGCAAAUCAAUGGAAGGACUUAAAACCACCUGAAUUAGGUGCCUAAGGC-CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUU ..((((((.....((((.(((.........((((((...)))))).....(((-......)))..))).)))).)))))).....(((((((((((((....))))))..)))))))... ( -39.30) >consensus UGGCAGCAAAUCAAUGGAAGGACUUAAAACCACCUGAAUUAGGUGCCUAAGGC_CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUU .(((((((.....((((.(((.........((((((...)))))).....(((.......)))..))).))))....))).))))(((((((((((((....))))))..)))))))... (-36.63 = -36.97 + 0.33)



| Location | 18,069,571 – 18,069,691 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -35.73 |
| Energy contribution | -36.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069571 120 - 22224390 AAGUCGUCCUACCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGGACACAGCAGGGUGGCACUGGGAGCCUUAGGCACCUAAUUCAGGUGGUUUUAAGUCCAUCCAUUGAUUUGCUGCCA ...(((((((...(((........))).)))))))((((.(((((.((.....((((((.(((..(((((......((((.....)))))))))..)))))))))..)).))))))))). ( -41.20) >DroSec_CAF1 7311 119 - 1 AAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG-GCCUUAGGCACCUAAUUCAGGUGGUUUUAAGUCCUUCCAUUGAUUUGCUGCCA ...((((((((..(((........)))))))))))((((.(((((.....))..(((((....(((-((...((.(((((.....))))).))....))))).)))))....))))))). ( -40.20) >DroSim_CAF1 11873 119 - 1 AAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG-GCCUUAGGCACCUAAUUCAGGUGGUUUUAAGUCCUUCCAUUGAUUUGCUGCCA ...((((((((..(((........)))))))))))((((.(((((.....))..(((((....(((-((...((.(((((.....))))).))....))))).)))))....))))))). ( -40.20) >consensus AAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG_GCCUUAGGCACCUAAUUCAGGUGGUUUUAAGUCCUUCCAUUGAUUUGCUGCCA ...((((((((..(((........)))))))))))((((.((((..(((......))).((.((((...(((((((((((.....)))))..))))))....)))).))..)))))))). (-35.73 = -36.07 + 0.33)

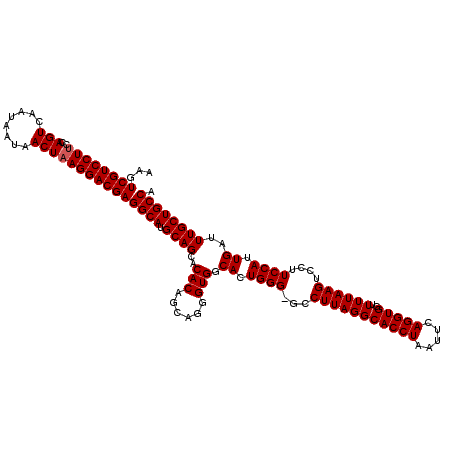

| Location | 18,069,611 – 18,069,731 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -41.13 |
| Energy contribution | -41.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069611 120 + 22224390 AGGUGCCUAAGGCUCCCAGUGCCACCCUGCUGUGUCCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGUAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGA .(((((.((((((...(((.(.(((......))).))))...)))(((((((((((((....))))))..))))))).....))).)))))(((((.((((.......))))..))))). ( -40.10) >DroSec_CAF1 7351 119 + 1 AGGUGCCUAAGGC-CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGA .(((((.((((((-..(((..(((......)).)..)))...)))(((((((((((((....))))))..))))))).....))).)))))(((((.((((.......))))..))))). ( -41.60) >DroSim_CAF1 11913 119 + 1 AGGUGCCUAAGGC-CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGA .(((((.((((((-..(((..(((......)).)..)))...)))(((((((((((((....))))))..))))))).....))).)))))(((((.((((.......))))..))))). ( -41.60) >consensus AGGUGCCUAAGGC_CCCAGUGCCACCCUGCUGUGUGCUGCAUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGA .(((((.((((((...(((..(((......)).)..)))...)))(((((((((((((....))))))..))))))).....))).)))))(((((.((((.......))))..))))). (-41.13 = -41.13 + -0.00)

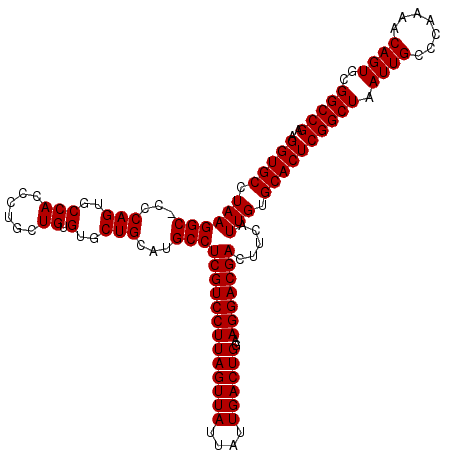

| Location | 18,069,611 – 18,069,731 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -41.33 |
| Energy contribution | -42.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069611 120 - 22224390 UCGGCCGCACUGUUUUGGGCAAUUAGCCGAGUGCACAAUGAAGUCGUCCUACCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGGACACAGCAGGGUGGCACUGGGAGCCUUAGGCACCU ...(((((.(((((.(((((.....)))...((((...((...(((((((...(((........))).)))))))..))))))...)).))))).)))))...(((.(((...))).))) ( -40.50) >DroSec_CAF1 7351 119 - 1 UCGGCCGCACUGUUUUGGGCAAUUAGCCGAGUGCACAAUGAAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG-GCCUUAGGCACCU ...(((((.(((((...(((.....)))..((((...(((...((((((((..(((........)))))))))))..)))...))))..))))).)))))...(((-(((...))).))) ( -44.50) >DroSim_CAF1 11913 119 - 1 UCGGCCGCACUGUUUUGGGCAAUUAGCCGAGUGCACAAUGAAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG-GCCUUAGGCACCU ...(((((.(((((...(((.....)))..((((...(((...((((((((..(((........)))))))))))..)))...))))..))))).)))))...(((-(((...))).))) ( -44.50) >consensus UCGGCCGCACUGUUUUGGGCAAUUAGCCGAGUGCACAAUGAAGUCGUCCUUCCAGUCAAUAAUAACUAAGGACGAGGCAUGCAGCACACAGCAGGGUGGCACUGGG_GCCUUAGGCACCU ...(((((.(((((...(((.....)))..((((...(((...((((((((..(((........)))))))))))..)))...))))..))))).)))))...(((.(((...))).))) (-41.33 = -42.00 + 0.67)


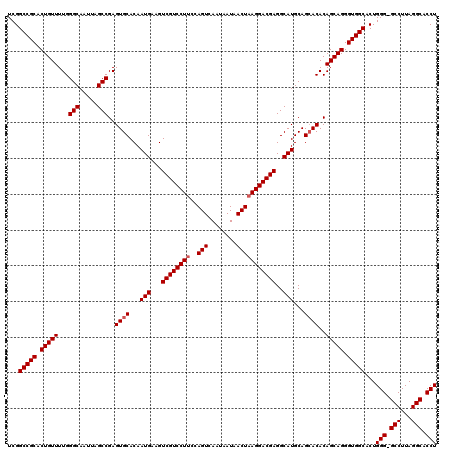
| Location | 18,069,651 – 18,069,771 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069651 120 + 22224390 AUGCCUCGUCCUUAGUUAUUAUUGACUGGUAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGAAACGCCAAUGAAAAUUGAUGCCUAAUUGCCCUAAGCCCGG ..((.(((((((((((((....))))))..))))))).(((((((.((..((((((.((((.......))))..))))))...)))))))))......................)).... ( -35.40) >DroSec_CAF1 7390 120 + 1 AUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGAAACGCCAAUGAAAAUUGAUGCCUAAUUGCCCUAAGCCCGG ..((.(((((((((((((....))))))..))))))).(((((((.((..((((((.((((.......))))..))))))...)))))))))......................)).... ( -35.40) >DroSim_CAF1 11952 120 + 1 AUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGAAACGCCAAUGAAAAUUGAUGCCUAAUUGCCCUAAGCCCGG ..((.(((((((((((((....))))))..))))))).(((((((.((..((((((.((((.......))))..))))))...)))))))))......................)).... ( -35.40) >consensus AUGCCUCGUCCUUAGUUAUUAUUGACUGGAAGGACGACUUCAUUGUGCACUCGGCUAAUUGCCCAAAACAGUGCGGCCGAAACGCCAAUGAAAAUUGAUGCCUAAUUGCCCUAAGCCCGG ..((.(((((((((((((....))))))..))))))).(((((((.((..((((((.((((.......))))..))))))...)))))))))......................)).... (-35.40 = -35.40 + -0.00)

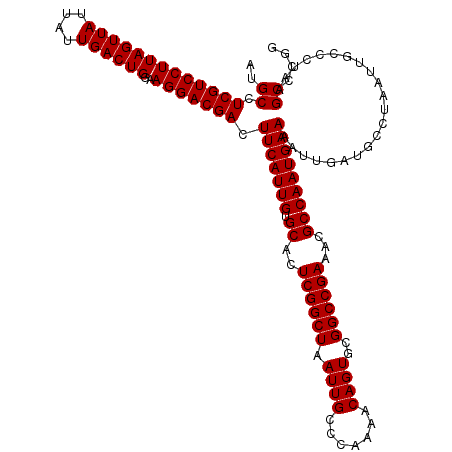
| Location | 18,069,731 – 18,069,851 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -47.23 |
| Consensus MFE | -45.37 |
| Energy contribution | -45.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18069731 120 - 22224390 ACCCCUUUGACUUGGCCUUUUGGGUGGCGGUGCCUUGGCUACGCUGCGCUCCGAGUGGAUUGCUCUGGCCGGGACAAUCGCCGGGCUUAGGGCAAUUAGGCAUCAAUUUUCAUUGGCGUU ((.((..(((....(((........)))(((((((......((((.((...))))))(((((((((((((.((.(....))).))).))))))))))))))))).....)))..)).)). ( -44.80) >DroSec_CAF1 7470 120 - 1 ACCCCUUUGACUUGGCCUUUUGGGUGGCGGUGCCUUGGCUACGCUCCGCUCCGAGUGGAUUGCUCUGGCCGGGACAAUCGCCGGGCUUAGGGCAAUUAGGCAUCAAUUUUCAUUGGCGUU ((.((..(((....(((........)))(((((((......(((((......)))))(((((((((((((.((.(....))).))).))))))))))))))))).....)))..)).)). ( -47.40) >DroSim_CAF1 12032 120 - 1 ACCCCUUUGACUUGGCCUUUUGGGUGGCGGUGCCUUGGCUACGCUCCGCUCCGAGUGGAUUGCUCUGGCCUGGACAAUCGCCGGGCUUAGGGCAAUUAGGCAUCAAUUUUCAUUGGCGUU ((.((..(((....(((........)))(((((((......(((((......)))))((((((((((((((((.(....))))))).))))))))))))))))).....)))..)).)). ( -49.50) >consensus ACCCCUUUGACUUGGCCUUUUGGGUGGCGGUGCCUUGGCUACGCUCCGCUCCGAGUGGAUUGCUCUGGCCGGGACAAUCGCCGGGCUUAGGGCAAUUAGGCAUCAAUUUUCAUUGGCGUU ((.((..(((....(((........)))(((((((......(((((......)))))(((((((((((((.((.(....))).))).))))))))))))))))).....)))..)).)). (-45.37 = -45.70 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:34 2006