
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,065,850 – 18,066,088 |
| Length | 238 |
| Max. P | 0.913741 |

| Location | 18,065,850 – 18,065,968 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -32.07 |
| Energy contribution | -31.19 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
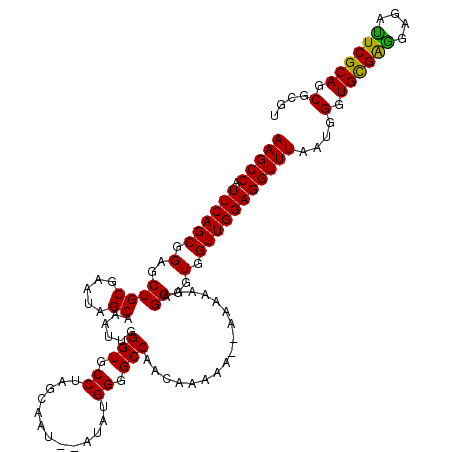
>X_DroMel_CAF1 18065850 118 + 22224390 AAGCCAUUCAGCGGAGCUGCGAAUAGCAAAUUGGGUGCCUAGCAAUGUAUAUGGGGCCAAUAGAAG--AAAAAAAGGGGUGGUUGGAGGUUUAACGGGUGUGAAGAGAUUCGCAGCGCGU (((((.((((((.(..((((.....))......(((.((.............)).)))........--.......))..).))))))))))).((((.((((((....)))))).).))) ( -30.72) >DroSec_CAF1 3686 115 + 1 AAGCCAUUCAGCGGAGCUGCGAAUAGCAAAUUGGGUGCCUAGCAAU--AUAUGGGGCCAACAAGAA---AAAAAAGGGGUGGUUGGAGGUUUAAUAGGUGCGGGGAGAUUCGCAGCGCGU ..........(((..(((((((((..(....(((....)))(((..--.(((..((((..(((...---.............)))..))))..)))..)))...)..))))))))).))) ( -30.39) >DroSim_CAF1 6823 118 + 1 AAGCCAUUCAGCGGAGCUGCGAAUAGCAAAUUGGGUGCCUAGCAAU--AUAUGGGGCCAACAAAAAAAAAAAAAAGGGGUGGUUGGAGGUUUAAUGGGUGCGAGGAGACUCGCAGCGCGU (((((.((((((.(..((((.....))......(((.((.......--....)).))).................))..).))))))))))).((((.((((((....)))))).).))) ( -36.50) >consensus AAGCCAUUCAGCGGAGCUGCGAAUAGCAAAUUGGGUGCCUAGCAAU__AUAUGGGGCCAACAAAAA__AAAAAAAGGGGUGGUUGGAGGUUUAAUGGGUGCGAGGAGAUUCGCAGCGCGU (((((.((((((.(..((((.....))......(((.((.............)).))).................))..).)))))))))))....(.((((((....)))))).).... (-32.07 = -31.19 + -0.88)


| Location | 18,065,850 – 18,065,968 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18065850 118 - 22224390 ACGCGCUGCGAAUCUCUUCACACCCGUUAAACCUCCAACCACCCCUUUUUUU--CUUCUAUUGGCCCCAUAUACAUUGCUAGGCACCCAAUUUGCUAUUCGCAGCUCCGCUGAAUGGCUU ....(((((((((.......................................--.......((((............))))((((.......)))))))))))))...(((....))).. ( -20.10) >DroSec_CAF1 3686 115 - 1 ACGCGCUGCGAAUCUCCCCGCACCUAUUAAACCUCCAACCACCCCUUUUUU---UUCUUGUUGGCCCCAUAU--AUUGCUAGGCACCCAAUUUGCUAUUCGCAGCUCCGCUGAAUGGCUU ....(((((((((......(((..(((.......(((((............---.....)))))......))--).)))..((((.......)))))))))))))...(((....))).. ( -23.65) >DroSim_CAF1 6823 118 - 1 ACGCGCUGCGAGUCUCCUCGCACCCAUUAAACCUCCAACCACCCCUUUUUUUUUUUUUUGUUGGCCCCAUAU--AUUGCUAGGCACCCAAUUUGCUAUUCGCAGCUCCGCUGAAUGGCUU ....((((((((....))))).............(((((....................)))))........--.......))).........(((((((((......)).))))))).. ( -23.55) >consensus ACGCGCUGCGAAUCUCCUCGCACCCAUUAAACCUCCAACCACCCCUUUUUUU__UUUUUGUUGGCCCCAUAU__AUUGCUAGGCACCCAAUUUGCUAUUCGCAGCUCCGCUGAAUGGCUU ....(((((((((......(((............(((((....................)))))............)))..((((.......)))))))))))))...(((....))).. (-20.65 = -21.10 + 0.45)



| Location | 18,065,890 – 18,066,008 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -27.01 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.831001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18065890 118 + 22224390 AGCAAUGUAUAUGGGGCCAAUAGAAG--AAAAAAAGGGGUGGUUGGAGGUUUAACGGGUGUGAAGAGAUUCGCAGCGCGUAAAGCGGCUCAAAGGACCCUAAUUAAAGUAAAUCAUGUUU ........((((((.(((....)...--.......(((((..((.(((((((.((((.((((((....)))))).).))).))))..))).))..))))).......))...)))))).. ( -24.90) >DroSec_CAF1 3726 115 + 1 AGCAAU--AUAUGGGGCCAACAAGAA---AAAAAAGGGGUGGUUGGAGGUUUAAUAGGUGCGGGGAGAUUCGCAGCGCGUAACGCGGCUCAAAGGAGCCUAAUUAAAGUAAAUCAUGUUU ......--((((((..(((((.....---............))))).(.((((((.(.((((((....)))))).)(((...)))(((((....)))))..)))))).)...)))))).. ( -27.23) >DroSim_CAF1 6863 118 + 1 AGCAAU--AUAUGGGGCCAACAAAAAAAAAAAAAAGGGGUGGUUGGAGGUUUAAUGGGUGCGAGGAGACUCGCAGCGCGUAUCGCGGCUCAAAGGAGCCUAAUUAAAGCAAAUCAUGUUU ......--((((((..(((((....................))))).(.((((((.(.((((((....)))))).)(((...)))(((((....)))))..)))))).)...)))))).. ( -35.85) >consensus AGCAAU__AUAUGGGGCCAACAAAAA__AAAAAAAGGGGUGGUUGGAGGUUUAAUGGGUGCGAGGAGAUUCGCAGCGCGUAACGCGGCUCAAAGGAGCCUAAUUAAAGUAAAUCAUGUUU ........((((((..(((((....................))))).(.((((((.(.((((((....)))))).)((.....))(((((....)))))..)))))).)...)))))).. (-27.01 = -26.35 + -0.66)



| Location | 18,065,968 – 18,066,088 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18065968 120 + 22224390 AAAGCGGCUCAAAGGACCCUAAUUAAAGUAAAUCAUGUUUUUCAUAACGCACCCUAAUUAUUUCACUAUUCAAAUCGAACUUUUCAAGUUCCACGUUGCCCGAAGUAAUUGGAUUAAGUA ...(((.....((((((....(((......)))...)))))).....)))...(((((((((((......(((...(((((.....)))))....)))...)))))))))))........ ( -17.90) >DroSec_CAF1 3801 120 + 1 AACGCGGCUCAAAGGAGCCUAAUUAAAGUAAAUCAUGUUUUUCAUAACGCACCCUAAUUACUUCACUAUUCAAAUCGAACUUUCCAAGUUCCACGUCGCCCGAAGUAAUUGGAUUAAGUA ...(((((((....))))).................(((......)))))...(((((((((((............(((((.....)))))...(.....))))))))))))........ ( -26.00) >DroSim_CAF1 6941 120 + 1 AUCGCGGCUCAAAGGAGCCUAAUUAAAGCAAAUCAUGUUUUUCAUAACGCACCCUAAUUACUUCACUAUUCAAAUCGAACUUUCCAAGUUCCACGUCGCCCGAAGUAAUUGGAUUAAGUA ...(((((((....))))).....((((((.....)))))).......))...(((((((((((............(((((.....)))))...(.....))))))))))))........ ( -27.30) >consensus AACGCGGCUCAAAGGAGCCUAAUUAAAGUAAAUCAUGUUUUUCAUAACGCACCCUAAUUACUUCACUAUUCAAAUCGAACUUUCCAAGUUCCACGUCGCCCGAAGUAAUUGGAUUAAGUA ...(((((((....))))).................(((......)))))...(((((((((((............(((((.....)))))...(.....))))))))))))........ (-23.25 = -23.37 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:24 2006