
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,060,455 – 18,060,615 |
| Length | 160 |
| Max. P | 0.882748 |

| Location | 18,060,455 – 18,060,575 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -38.93 |
| Energy contribution | -39.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060455 120 + 22224390 ACCAUAACCACCCACUUUGGCCGCCACCCAGAAAAGUUGGCAGCUGAAGGCGGCAGUAGCAGCCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUAC .......((.....(((..((.((((.(.......).)))).))..)))..((((((((((((((....))...)))))))))...........((((.....))))...)))))..... ( -40.20) >DroSec_CAF1 5622 120 + 1 ACCAUAACCACCCACUUUGGCCGCCACCCAGAAAAGUUGGCAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUAC ...............((((((.((((.(.......).))))....(((((((((((((....)))))))(((((((....))))))).................)))))))))))).... ( -41.20) >DroSim_CAF1 13848 120 + 1 ACCAUAACCACCCACUUUGGCCGCCACCCAGAAAAGUUGGCAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCGUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUAC ...............((((((.((((.(.......).))))....(((((((((((((....)))))))(((((((....))))))).................)))))))))))).... ( -41.70) >consensus ACCAUAACCACCCACUUUGGCCGCCACCCAGAAAAGUUGGCAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUAC ...............((((((.((((.(.......).))))....((((((((((((......))))))(((((((....))))))).................)))))))))))).... (-38.93 = -39.27 + 0.33)


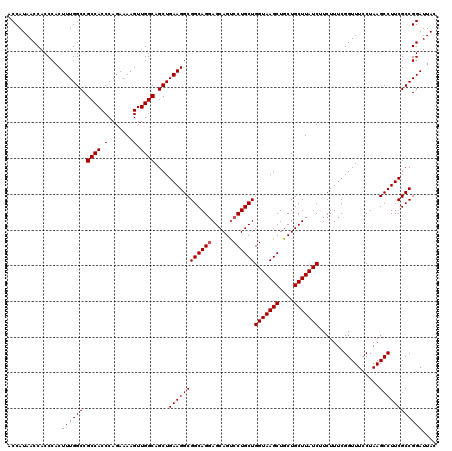
| Location | 18,060,455 – 18,060,575 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -45.47 |
| Consensus MFE | -43.89 |
| Energy contribution | -44.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060455 120 - 22224390 GUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGGCUGCUACUGCCGCCUUCAGCUGCCAACUUUUCUGGGUGGCGGCCAAAGUGGGUGGUUAUGGU ....((((((....)))..))).((((..........((((((.(((....)))...))))))...(((((((...(((((((.(((....))))))))))......)))))))..)))) ( -44.60) >DroSec_CAF1 5622 120 - 1 GUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUGCCAACUUUUCUGGGUGGCGGCCAAAGUGGGUGGUUAUGGU (((((((.((((((((...((....(....)....(((((....))))))).((((((....)))))).)))))).(((((((.(((....))))))))))....)).))..)))))... ( -45.90) >DroSim_CAF1 13848 120 - 1 GUAAUCCGGCGAAGGCUUAGGAAACCGAAAGACGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUGCCAACUUUUCUGGGUGGCGGCCAAAGUGGGUGGUUAUGGU (((((((.((((((((...((....(....)....(((((....))))))).((((((....)))))).)))))).(((((((.(((....))))))))))....)).))..)))))... ( -45.90) >consensus GUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUGCCAACUUUUCUGGGUGGCGGCCAAAGUGGGUGGUUAUGGU (((((((.((((((((...((....(....)....(((((....))))))).((((((....)))))).)))))).(((((((.(((....))))))))))....)).))..)))))... (-43.89 = -44.00 + 0.11)

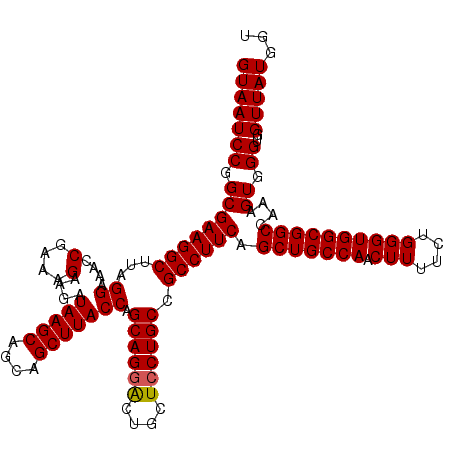
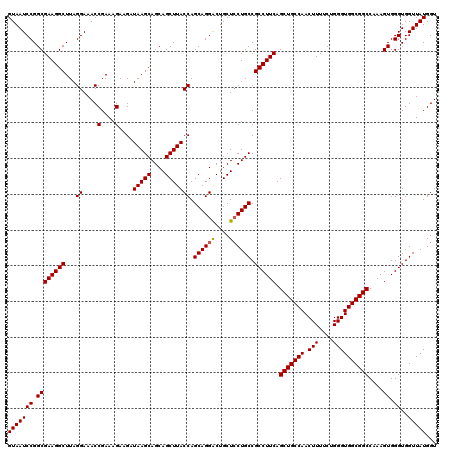
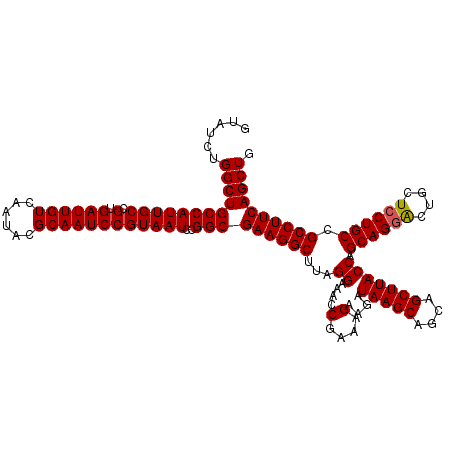
| Location | 18,060,495 – 18,060,615 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -42.53 |
| Energy contribution | -42.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060495 120 + 22224390 CAGCUGAAGGCGGCAGUAGCAGCCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUACGGAUUGCGUAUUGACAAUCACGGCAAUGGCAGCCAGAUAC ..((((((((((((((((((.(((.....)))..)))))))))...........((...))...))))))((((.....(((((((........))))).))....)))))))....... ( -41.60) >DroSec_CAF1 5662 120 + 1 CAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUACGGAUUGCGUAUUGACAAUCACGGCAAUGGCAGCCAGAUAC ..((((((((((((((((....)))))))(((((((....))))))).................))))))((((.....(((((((........))))).))....)))))))....... ( -44.80) >DroSim_CAF1 13888 120 + 1 CAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCGUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUACGGAUUGCGUAUUGACAAUCACGGCAAUGGCAGCCAGAUAC ..((((((((((((((((....)))))))(((((((....))))))).................))))))((((.....(((((((........))))).))....)))))))....... ( -45.30) >consensus CAGCUGAAGGCGGCAGGAGCAGUCCUGCUGGUAAGCUGCUGCUUAUCUUCUUUCGGUUUCCUAAGCCUUCGCCGGAUUACGGAUUGCGUAUUGACAAUCACGGCAAUGGCAGCCAGAUAC ..(((((((((((((((......))))))(((((((....))))))).................))))))((((.....(((((((........))))).))....)))))))....... (-42.53 = -42.87 + 0.33)


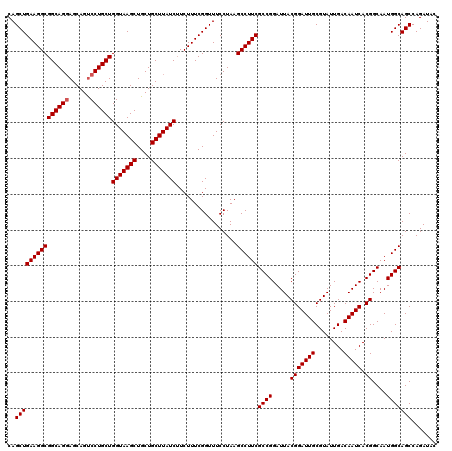
| Location | 18,060,495 – 18,060,615 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -41.29 |
| Energy contribution | -41.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060495 120 - 22224390 GUAUCUGGCUGCCAUUGCCGUGAUUGUCAAUACGCAAUCCGUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGGCUGCUACUGCCGCCUUCAGCUG (((...((((((..((((((.(((......((((.....)))))))))))))..((((((....)(....)....))))).)))))))))(((((((((.((....)).)))))..)))) ( -41.20) >DroSec_CAF1 5662 120 - 1 GUAUCUGGCUGCCAUUGCCGUGAUUGUCAAUACGCAAUCCGUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUG ......((((((((((((.(.((((((......))))))))))))..)))((((((...((....(....)....(((((....))))))).((((((....)))))).)))))))))). ( -43.30) >DroSim_CAF1 13888 120 - 1 GUAUCUGGCUGCCAUUGCCGUGAUUGUCAAUACGCAAUCCGUAAUCCGGCGAAGGCUUAGGAAACCGAAAGACGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUG ......((((((((((((.(.((((((......))))))))))))..)))((((((...((....(....)....(((((....))))))).((((((....)))))).)))))))))). ( -43.30) >consensus GUAUCUGGCUGCCAUUGCCGUGAUUGUCAAUACGCAAUCCGUAAUCCGGCGAAGGCUUAGGAAACCGAAAGAAGAUAAGCAGCAGCUUACCAGCAGGACUGCUCCUGCCGCCUUCAGCUG ......((((((((((((.(.((((((......))))))))))))..)))((((((...((....(....)....(((((....))))))).((((((....)))))).)))))))))). (-41.29 = -41.40 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:08 2006