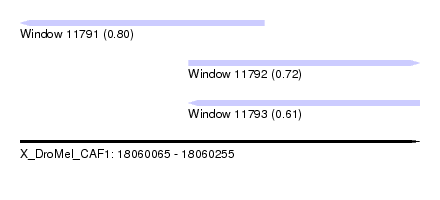
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,060,065 – 18,060,255 |
| Length | 190 |
| Max. P | 0.798271 |

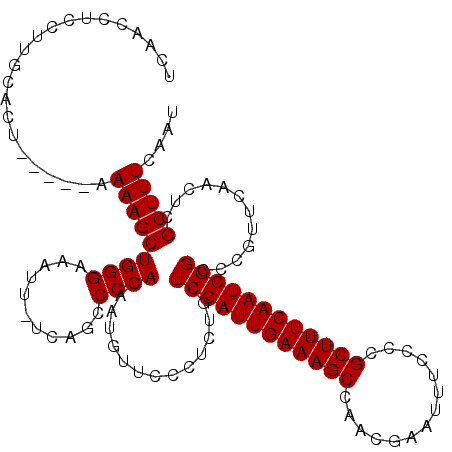
| Location | 18,060,065 – 18,060,181 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.21 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060065 116 - 22224390 UCAACCUCCUUGCACUU----UAAAGCUGGGAAAUUUUUAGCCCCAAAAUGUUCCCUCUAUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCGCGUUCAACUCGCUUUCAAU ...........((....----.......(((((..((((......))))..)))))......(((((((((((..............)))))))))))))(((.......)))....... ( -25.64) >DroSec_CAF1 5241 119 - 1 UCAACCUCCUUGCACUAUGGUAAAAGCUGGGAAAUU-UCAGCCCCACAAUGUUCCCUCUGUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACUCGCUUUCAAU ...(((............))).(((((.(((((...-..............)))))...(..(((((((((((..............)))))))))))..)..........))))).... ( -24.67) >DroSim_CAF1 13470 114 - 1 UCAACCUCCUUGCACU-----AAAAGCUGGGAAAUU-UCAGCCCCACAAUGUUGCCUCUGUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACUCGCUUUCAAU ................-----.(((((((((.....-.....))))....((((...(.(..(((((((((((..............)))))))))))..).)..))))..))))).... ( -26.44) >consensus UCAACCUCCUUGCACU_____AAAAGCUGGGAAAUU_UCAGCCCCACAAUGUUCCCUCUGUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACUCGCUUUCAAU ......................(((((((((...........))))...............((((((((((((..............))))))))))))............))))).... (-22.21 = -22.21 + -0.00)


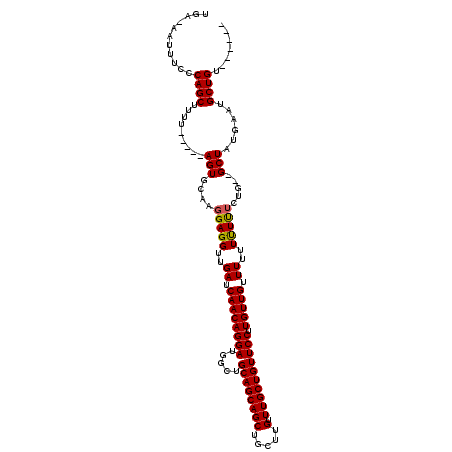
| Location | 18,060,145 – 18,060,255 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.89 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060145 110 + 22224390 UAAAAAUUUCCCAGCUUUA----AAGUGCAAGGAGGUUGAUCAACAGGAUGGCUGCAGCAGCUGCUUGUUUGCUGUUCCUUGUUGUUUUUUCUU------GCUGUGAAUGCUGUUAUUAU ...........((((((((----....(((((((((.....((((((((((((.(((((....)).)))..)))).))).)))))..)))))))------))..)))).))))....... ( -32.10) >DroSec_CAF1 5321 111 + 1 UGA-AAUUUCCCAGCUUUUACCAUAGUGCAAGGAGGUUGAUCAACAGGAAGGCUGCAGCAGCUGCUUGUUUGCUGUUCCUUGUUGUUUUUUUUUUCUG--GCUAUGAAUGCUGU------ ...-.......((((......((((((...((((((..((..(((((.((((..((((((((.....).))))))).)))).)))))))..)))))).--))))))...)))).------ ( -32.40) >DroSim_CAF1 13550 108 + 1 UGA-AAUUUCCCAGCUUUU-----AGUGCAAGGAGGUUGAUCAACAGGAUGGCUGCAGCAGCUGCUUGUUUGCUGUUCCUUGUUGUUUUUUUUCUCUGAAGCUAUGAAUGCUGU------ ...-.......((((.(((-----(..((..(((((..((.((((((((((((.(((((....)).)))..)))).))).)))))...))..)))))...))..)))).)))).------ ( -31.50) >consensus UGA_AAUUUCCCAGCUUUU_____AGUGCAAGGAGGUUGAUCAACAGGAUGGCUGCAGCAGCUGCUUGUUUGCUGUUCCUUGUUGUUUUUUUUUUCUG__GCUAUGAAUGCUGU______ ...........((((.........(((....(((((..((.((((((((.....((((((((.....).)))))))))).))))).))..))))).....)))......))))....... (-24.00 = -23.89 + -0.11)


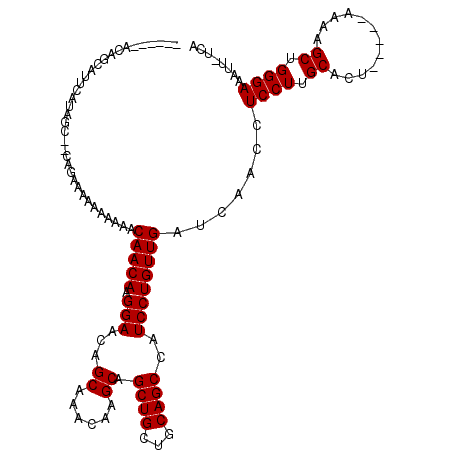
| Location | 18,060,145 – 18,060,255 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18060145 110 - 22224390 AUAAUAACAGCAUUCACAGC------AAGAAAAAACAACAAGGAACAGCAAACAAGCAGCUGCUGCAGCCAUCCUGUUGAUCAACCUCCUUGCACUU----UAAAGCUGGGAAAUUUUUA .......((((.......((------(((......(((((.(((...((......)).((((...))))..)))))))).........)))))....----....))))........... ( -21.62) >DroSec_CAF1 5321 111 - 1 ------ACAGCAUUCAUAGC--CAGAAAAAAAAAACAACAAGGAACAGCAAACAAGCAGCUGCUGCAGCCUUCCUGUUGAUCAACCUCCUUGCACUAUGGUAAAAGCUGGGAAAUU-UCA ------.((((..((((((.--(((..........(((((.((((..((......)).((((...)))).)))))))))..........)))..)))))).....)))).......-... ( -22.35) >DroSim_CAF1 13550 108 - 1 ------ACAGCAUUCAUAGCUUCAGAGAAAAAAAACAACAAGGAACAGCAAACAAGCAGCUGCUGCAGCCAUCCUGUUGAUCAACCUCCUUGCACU-----AAAAGCUGGGAAAUU-UCA ------..(((.......)))...((((.......(((((.(((...((......)).((((...))))..)))))))).......((((.((...-----....)).))))..))-)). ( -22.50) >consensus ______ACAGCAUUCAUAGC__CAGAAAAAAAAAACAACAAGGAACAGCAAACAAGCAGCUGCUGCAGCCAUCCUGUUGAUCAACCUCCUUGCACU_____AAAAGCUGGGAAAUU_UCA ...................................(((((.(((...((......)).((((...))))..)))))))).......((((.((............)).))))........ (-18.47 = -18.47 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:05 2006