
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,057,734 – 18,057,932 |
| Length | 198 |
| Max. P | 0.979079 |

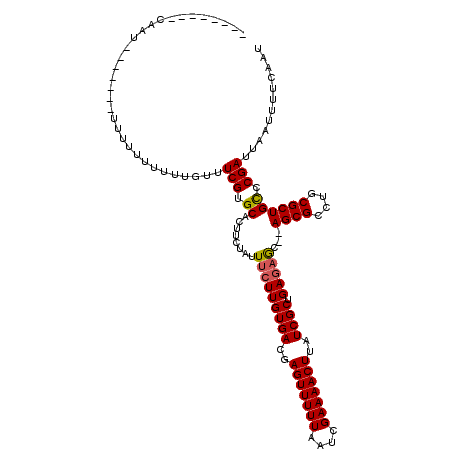
| Location | 18,057,734 – 18,057,852 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18057734 118 + 22224390 CUGUGCCACAUUUUUUGUUUUUUUUUUUUCAUUCGUGCACUUCUAUUUCUUGUGACGAGUUUUUAAUCGAAAACUUAUCGCUGAGAGC--AGCGCCUGCGCUGCCCGAUUAAUUUUCAAU ..((((.((.........................)))))).......((((((((..(((((((....)))))))..)))).))))((--((((....))))))................ ( -28.11) >DroSec_CAF1 2358 104 + 1 --------CAAU-------UUUU-UUUUUGUUUCGUGCUCUUCUAUUGCUUGUGACGAGUUUUUAAUCGAAAACUUAUCGCUGAUAACUGAGCGCCUGCGCUGUCCGAUUAAUUUUCAAU --------....-------....-.....((...((((((...((((....((((..(((((((....)))))))..)))).))))...))))))..))..................... ( -21.70) >DroSim_CAF1 2375 103 + 1 --------CAAU-------UUUUUUUUUUGUUUCGUGCACUUCUAUUUCUUGUGACGAGUUUUUAAUCGAAAACUUAUCGCUGAGAGC--AGCGCCUGCGCUGCCCGAUUAAUUUUCAAU --------....-------.............(((............((((((((..(((((((....)))))))..)))).))))((--((((....)))))).)))............ ( -26.10) >consensus ________CAAU_______UUUUUUUUUUGUUUCGUGCACUUCUAUUUCUUGUGACGAGUUUUUAAUCGAAAACUUAUCGCUGAGAGC__AGCGCCUGCGCUGCCCGAUUAAUUUUCAAU ................................(((.((........(((((((((..(((((((....)))))))..)))).)))))...((((....)))))).)))............ (-21.34 = -21.57 + 0.23)


| Location | 18,057,734 – 18,057,852 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.84 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18057734 118 - 22224390 AUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAAGUGCACGAAUGAAAAAAAAAAAACAAAAAAUGUGGCACAG ((((((((((.(((((((((((....))))--))).....)))).)))))))))).......(((((((...........))).))))................................ ( -27.70) >DroSec_CAF1 2358 104 - 1 AUUGAAAAUUAAUCGGACAGCGCAGGCGCUCAGUUAUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGCAAUAGAAGAGCACGAAACAAAAA-AAAA-------AUUG-------- ............(((....((....))((((..((((..((((..((((((......))))))..))....)).))))..)))).)))........-....-------....-------- ( -17.20) >DroSim_CAF1 2375 103 - 1 AUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAAGUGCACGAAACAAAAAAAAAA-------AUUG-------- ((((((((((.(((((((((((....))))--))).....)))).)))))))))).......(((((((...........))).)))).............-------....-------- ( -27.70) >consensus AUUGAAAAUUAAUCGGGCAGCGCAGGCGCU__GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAAGUGCACGAAACAAAAAAAAAA_______AUUG________ ((((((((((.(((((((((((....))))..))).....)))).)))))))))).......(((((((...........))).))))................................ (-20.31 = -20.20 + -0.11)



| Location | 18,057,774 – 18,057,892 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -31.61 |
| Energy contribution | -31.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18057774 118 - 22224390 AACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAA ..((((..((((((((((((.(.(((....)))(((((.......)))))....).))))))).))))).--((.....))((..((((((......))))))..)).))))........ ( -32.10) >DroSec_CAF1 2382 120 - 1 AACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGACAGCGCAGGCGCUCAGUUAUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGCAAUAGAA ...(((((((((((((((((.(.(((....)))(((((.......)))))....).))))))).)))))..........(.((..((((((......))))))..)).).)))))..... ( -32.90) >DroSim_CAF1 2400 118 - 1 AACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAA ..((((..((((((((((((.(.(((....)))(((((.......)))))....).))))))).))))).--((.....))((..((((((......))))))..)).))))........ ( -32.10) >consensus AACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU__GCUCUCAGCGAUAAGUUUUCGAUUAAAAACUCGUCACAAGAAAUAGAA ..((((..((((((((((((.(.(((....)))(((((.......)))))....).))))))).)))))...((.....))((..((((((......))))))..)).))))........ (-31.61 = -31.17 + -0.44)



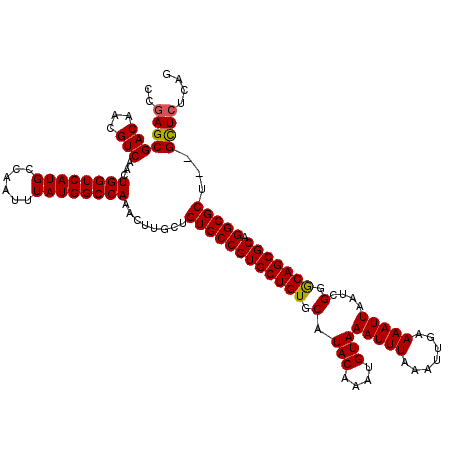
| Location | 18,057,814 – 18,057,932 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -35.61 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18057814 118 - 22224390 CCGAGCGACAACGUCAACUGGUCAUGCCAAUUUAUGGCCAAACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAG ..(((((((...)))...((((((((......))))))))........((((((((((((.(.(((....)))(((((.......)))))....).))))))).))))).--)))).... ( -37.80) >DroSec_CAF1 2422 120 - 1 CCGAGCGACAACGUCAACUGGUCAUGCCAAUUUAUGGCCAAACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGACAGCGCAGGCGCUCAGUUAUCAG ..((((....(((.(((.((((((((......))))))))...))).)))((((((((((.(.(((....)))(((((.......)))))....).))))))).)))))))......... ( -35.90) >DroSim_CAF1 2440 118 - 1 CCGAGCGACAACGUCAACUGGUCAUGCCAAUUUAUGGCCAAACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU--GCUCUCAG ..(((((((...)))...((((((((......))))))))........((((((((((((.(.(((....)))(((((.......)))))....).))))))).))))).--)))).... ( -37.80) >consensus CCGAGCGACAACGUCAACUGGUCAUGCCAAUUUAUGGCCAAACUUGCUGUGCCGUGCUGUGCAUACAAAUGUAAAUUUAAAUUGAAAAUUAAUCGGGCAGCGCAGGCGCU__GCUCUCAG ..(((((((...)))...((((((((......))))))))........((((((((((((.(.(((....)))(((((.......)))))....).))))))).)))))...)))).... (-35.61 = -35.50 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:01 2006