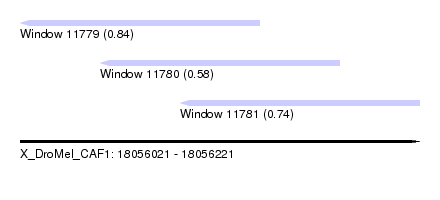
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,056,021 – 18,056,221 |
| Length | 200 |
| Max. P | 0.837262 |

| Location | 18,056,021 – 18,056,141 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18056021 120 - 22224390 CCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAACAAGCGUGAAGUGAAGCAGCAACGUAAAAUGUAAAACAAA .(((((((((((...((((((((....))))))))...........(((....))).....................)))))))))))..((...(((...........)))...))... ( -22.60) >DroSec_CAF1 718 120 - 1 CCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAACAAGCGUGAAGGGUAGCAGCAACGUAAAAUGUAAAACAAA .(((((((((((...((((((((....))))))))...........(((....))).....................)))))))))))..(..(.(((...........))).)..)... ( -22.10) >DroSim_CAF1 717 120 - 1 CCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAACAAGCGUGAAGGGUAGCAGCAGCGUAAAAUGUAAAACAAA .(((((((((((...((((((((....))))))))...........(((....))).....................))))))))))).......((....))................. ( -22.90) >consensus CCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAACAAGCGUGAAGGGUAGCAGCAACGUAAAAUGUAAAACAAA .(((((((((((...((((((((....))))))))...........(((....))).....................)))))))))))..........(((........)))........ (-21.77 = -21.77 + 0.00)



| Location | 18,056,061 – 18,056,181 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18056061 120 - 22224390 UACCAUACCCCCAUUGCUGCCGAAUCAUCUGGCUACCCCCCCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAA ................((((.((....))(((((......((((.(((((((...((((((((....))))))))...)))))))))))..))))).))))................... ( -23.80) >DroSec_CAF1 758 103 - 1 UACCGUUCCCC---------------GUCUGGCCACC--ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAA ...........---------------(..((((....--.((((.(((((((...((((((((....))))))))...)))))))))))...))))..)..................... ( -22.30) >DroSim_CAF1 757 103 - 1 UACCGUUCCCC---------------GUCUGGCCACC--ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAA ...........---------------(..((((....--.((((.(((((((...((((((((....))))))))...)))))))))))...))))..)..................... ( -22.30) >consensus UACCGUUCCCC_______________GUCUGGCCACC__ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGAAUAAAAUGGAAAGCCAAGCAGAAACUACCGAAAAAAGAAA ..........................((.((((.......((((.(((((((...((((((((....))))))))...)))))))))))...)))).))..................... (-18.50 = -18.83 + 0.33)



| Location | 18,056,101 – 18,056,221 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -25.31 |
| Energy contribution | -24.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18056101 120 - 22224390 GGGAGGAUAAACAGGCCAAGUCACUCUCACAUGGCAUUCCUACCAUACCCCCAUUGCUGCCGAAUCAUCUGGCUACCCCCCCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGA ((...(((((((((((((.((.......)).)))).......................((((.......))))...........(((((((.....)))))))..)))))))))..)).. ( -28.30) >DroSec_CAF1 798 102 - 1 GGG-GGAUAAACAGGCCAAGUCACUCUCACAUGGCCUUCCUACCGUUCCCC---------------GUCUGGCCACC--ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGA (((-((((....((((((.((.......)).)))))).......)))))))---------------(..((((...(--(.((.(((((((.....))))))))).))...))))..).. ( -29.30) >DroSim_CAF1 797 103 - 1 GGGAGGAUAAACAGGCCAAGUCACUCUCACAUGGCCUUCCUACCGUUCCCC---------------GUCUGGCCACC--ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGA ((.((((.....((((((.((.......)).)))))))))).))......(---------------(..((((...(--(.((.(((((((.....))))))))).))...))))..)). ( -27.50) >consensus GGGAGGAUAAACAGGCCAAGUCACUCUCACAUGGCCUUCCUACCGUUCCCC_______________GUCUGGCCACC__ACCAUGUUUGUUUAAAUGACAAACUGCUGUUUGUUAACCGA ((...(((((((((((((.((.......))((((........)))).......................)))))..........(((((((.....)))))))...))))))))..)).. (-25.31 = -24.87 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:53 2006