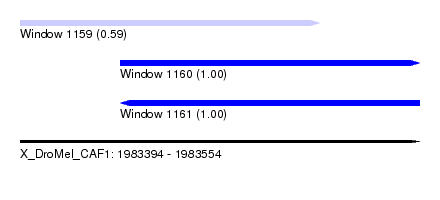
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,983,394 – 1,983,554 |
| Length | 160 |
| Max. P | 0.996140 |

| Location | 1,983,394 – 1,983,514 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -20.99 |
| Energy contribution | -22.30 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1983394 120 + 22224390 AAAAUAGGAAACGAUUCUGAAAGCUUUGGGGUGGCCUCUGUCUGCAUCUUCUCCUCAGCCAUGGCUUCAUCCAGCCUGUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCACCUGUC ...(((((...((....))......(((((((((.(..((....))...........((((((((......................))))))))).)))))))))........))))). ( -33.95) >DroSec_CAF1 16841 120 + 1 AAAAUAGGAAAUGAUUCUGAAAGCUUUGGGGUGGCCUCCGUCUGCAUCUUUUCCUCAGCCAUGGCUUCAUCCAGUCUGUCCACCAAUGCCAUGGCAUCCACCCCAACUAUACCGCCUGUC ...(((((.........(....)..(((((((((.....(....)............((((((((......................))))))))..)))))))))........))))). ( -32.15) >DroEre_CAF1 6527 120 + 1 UGAAUUGGCAAAGCCUCGGAUAUUGUCUGAAAUGAUUCUGUCUGCAUUUUUUCCUCGGCCAUAGCUUCAUCCAGUCUAUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCGCCUGUC .....(((((......((((((..(((......)))..))))))............((..((((((......)).))))...))..))))).((((................)))).... ( -22.29) >DroYak_CAF1 17274 120 + 1 AAAACAGAAAACGAUUCUAAAAGCUUCGGGGUGGUCUCCGUCUGCAUCUUCUCCUCAGCCAUGGCUUCGUCCAGUUUGUCCACCAAUGCCAUGGCGUCUACCCCAACUAUACCGCCUGUU ..(((((.....((..(.....)..))(((((((.(...(....)............((((((((......................))))))))).)))))))...........))))) ( -32.25) >consensus AAAAUAGGAAACGAUUCUGAAAGCUUCGGGGUGGCCUCCGUCUGCAUCUUCUCCUCAGCCAUGGCUUCAUCCAGUCUGUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCGCCUGUC ....(((((.....)))))........(((((((.....(....)............((((((((......................))))))))..)))))))................ (-20.99 = -22.30 + 1.31)

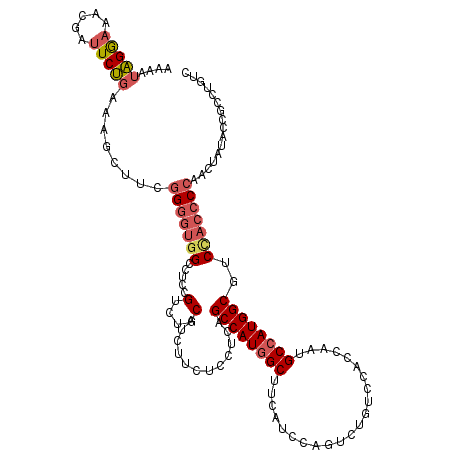
| Location | 1,983,434 – 1,983,554 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -27.11 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1983434 120 + 22224390 UCUGCAUCUUCUCCUCAGCCAUGGCUUCAUCCAGCCUGUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCACCUGUCCCCAGGCCGUUCGUCUCUCCUGAUCCAACUCCACCGAUGC ...(((((.........((((((((......................))))))))...................((((....))))..(((.(..((....))..))))......))))) ( -25.65) >DroSec_CAF1 16881 120 + 1 UCUGCAUCUUUUCCUCAGCCAUGGCUUCAUCCAGUCUGUCCACCAAUGCCAUGGCAUCCACCCCAACUAUACCGCCUGUCCCCAGGCCGUUGGUCUCUCCUGAUCCAACUCCACCGAUGC ...(((((.........((((((((......................))))))))..................(((((....))))).(((((((......)).)))))......))))) ( -35.55) >DroEre_CAF1 6567 120 + 1 UCUGCAUUUUUUCCUCGGCCAUAGCUUCAUCCAGUCUAUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCGCCUGUCCCCAGGCCGUUGGUCUCUCCUGCUCCACCUCCUCCGAUGC ...(((((.........(((((.((......................)).))))).......(((((......(((((....))))).)))))......................))))) ( -22.75) >DroYak_CAF1 17314 120 + 1 UCUGCAUCUUCUCCUCAGCCAUGGCUUCGUCCAGUUUGUCCACCAAUGCCAUGGCGUCUACCCCAACUAUACCGCCUGUUCCCAGGCCGUUGGUCUCUGCUGUUCCACCUCCUCCGAUGC ...(((((.........((((((((......................))))))))((..(..(((((......(((((....))))).)))))..)..))...............))))) ( -32.45) >consensus UCUGCAUCUUCUCCUCAGCCAUGGCUUCAUCCAGUCUGUCCACCAAUGCCAUGGCGUCCACCCCAACUAUACCGCCUGUCCCCAGGCCGUUGGUCUCUCCUGAUCCAACUCCACCGAUGC ...(((((.........((((((((......................))))))))..........((((.((.(((((....))))).)))))).....................))))) (-27.11 = -27.68 + 0.56)

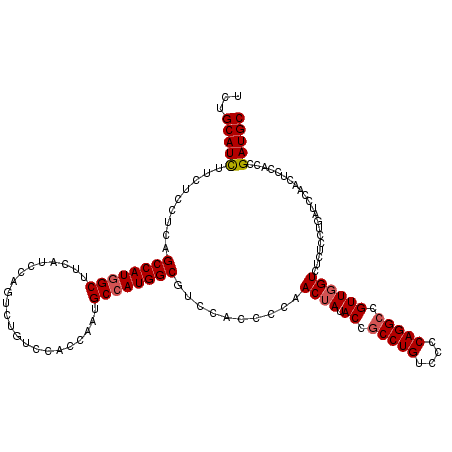
| Location | 1,983,434 – 1,983,554 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -38.04 |
| Energy contribution | -37.98 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
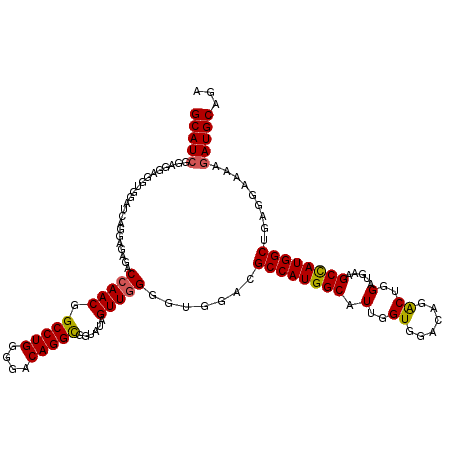
>X_DroMel_CAF1 1983434 120 - 22224390 GCAUCGGUGGAGUUGGAUCAGGAGAGACGAACGGCCUGGGGACAGGUGGUAUAGUUGGGGUGGACGCCAUGGCAUUGGUGGACAGGCUGGAUGAAGCCAUGGCUGAGGAGAAGAUGCAGA (((((.......(((..((....))..)))((.(((((....))))).)).(((((....((..(((((......)))))..))((((......))))..))))).......)))))... ( -43.10) >DroSec_CAF1 16881 120 - 1 GCAUCGGUGGAGUUGGAUCAGGAGAGACCAACGGCCUGGGGACAGGCGGUAUAGUUGGGGUGGAUGCCAUGGCAUUGGUGGACAGACUGGAUGAAGCCAUGGCUGAGGAAAAGAUGCAGA (((((......(((((.((......))))))).(((((....)))))..................((((((((.(..((......))..).....)))))))).........)))))... ( -43.50) >DroEre_CAF1 6567 120 - 1 GCAUCGGAGGAGGUGGAGCAGGAGAGACCAACGGCCUGGGGACAGGCGGUAUAGUUGGGGUGGACGCCAUGGCAUUGGUGGAUAGACUGGAUGAAGCUAUGGCCGAGGAAAAAAUGCAGA ((((.......(((.((((.((.....)).((.(((((....))))).))....(..(..((..(((((......)))))..))..)..).....))).).))).........))))... ( -37.99) >DroYak_CAF1 17314 120 - 1 GCAUCGGAGGAGGUGGAACAGCAGAGACCAACGGCCUGGGAACAGGCGGUAUAGUUGGGGUAGACGCCAUGGCAUUGGUGGACAAACUGGACGAAGCCAUGGCUGAGGAGAAGAUGCAGA (((((......................(((((.(((((....)))))......))))).......((((((((.(((..(......)....))).)))))))).........)))))... ( -41.50) >consensus GCAUCGGAGGAGGUGGAUCAGGAGAGACCAACGGCCUGGGGACAGGCGGUAUAGUUGGGGUGGACGCCAUGGCAUUGGUGGACAGACUGGAUGAAGCCAUGGCUGAGGAAAAGAUGCAGA (((((......................(((((.(((((....)))))......))))).......((((((((.(..((......))..).....)))))))).........)))))... (-38.04 = -37.98 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:03 2006