
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,031,826 – 18,032,061 |
| Length | 235 |
| Max. P | 0.730629 |

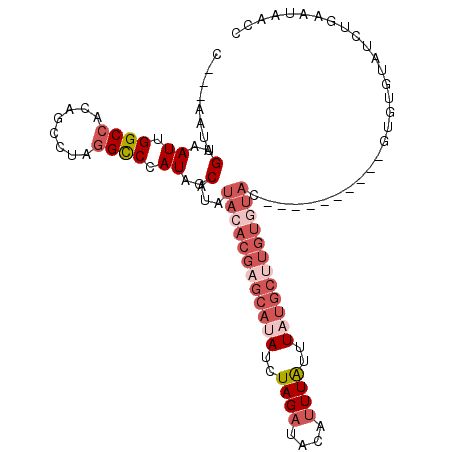
| Location | 18,031,826 – 18,031,924 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -12.98 |
| Energy contribution | -15.78 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18031826 98 - 22224390 CAGCAAUGGAAAUUGGCCACAGCCUAGGCCCAUAGCAUAUACACGAGCAUAUCUAGAUACAUUUAUUUAUGCCUGUGUAC----------GUGUGUAUCUGAAUAACC ..((.((((.....(((....))).....)))).))(((((((((.(((((..((((((....))))))....))))).)----------)))))))).......... ( -26.30) >DroSec_CAF1 19811 95 - 1 C---AAUUGAAAUUGGCCACAGCCUAGGUCCAUAGCAUAUACACGAGCAUAUCUAGAUACAUUUAUUUAUGCUUGUGUAC----------GUGUGUAUCUGAAUAAUC .---..........(((....)))(((((.....(((..((((((((((((..((((....))))..)))))))))))).----------.)))..)))))....... ( -27.20) >DroSim_CAF1 20140 95 - 1 C---AAUUGAAAUUGGCCACAGCCUAGGCCCAUAGCAUAUACACGAGCAUAUCUAGAUACAUUUAUUUAUGCUUGUGUAC----------GUGUGUAUCUGAAUAACC .---..........((((........))))....(((..((((((((((((..((((....))))..)))))))))))).----------.))).............. ( -28.70) >DroEre_CAF1 21751 95 - 1 C---AAUUGAAAUUGGCCGCAGCCUAUGUCCAUGGCAUAUAGACGAGCACAUCUAGAUACACUUGUUUAUGCUUGUGUAC----------GUGUGUAUCUGAAUAACC .---..(((.....(((....)))(((((.....)))))....))).......(((((((((.(((.(((....))).))----------).)))))))))....... ( -20.00) >DroYak_CAF1 19627 93 - 1 C---AAUUGAAAUUGGCCGCAGCGUAGGCCCAUGGC------------ACAUCUAGAUACAUUUAUGUACGCUUGUGUACGUGUACGUACGUGUGUAUCUGAAUAACC .---...((..((.((((........)))).))..)------------)....(((((((((.(((((((((........)))))))))...)))))))))....... ( -29.30) >consensus C___AAUUGAAAUUGGCCACAGCCUAGGCCCAUAGCAUAUACACGAGCAUAUCUAGAUACAUUUAUUUAUGCUUGUGUAC__________GUGUGUAUCUGAAUAACC ........(..((.((((........)))).))..)...((((((((((((..((((....))))..))))))))))))............................. (-12.98 = -15.78 + 2.80)


| Location | 18,031,924 – 18,032,031 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -22.46 |
| Energy contribution | -24.24 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18031924 107 + 22224390 CGUGAAGCAGCUGAAGCUGUAUUUGGCGCCCAACGUGGGGCACAUUCAACCGGAAUAAAAAGGUUCACCUAUUGGAGGGCCCUUGCCCAAACGCCCCAUCACCAGAC--------- .((((.(((((....)))))..((((...))))..((((((...(((((...((((......)))).....)))))((((....))))....)))))))))).....--------- ( -34.50) >DroSec_CAF1 19906 107 + 1 CGUGCAGCAGCUGAAGCUGAAUUUGGCGCCCAACAUGGGGCAUAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCCUUGCCCAAACGCCCCAUCAUCAGAC--------- .((((..((((....))))......)))).....(((((((.(((((.....)))))....(((((.((....)).)))))...........)))))))........--------- ( -34.90) >DroSim_CAF1 20235 107 + 1 CGUGCAGCAGCUGAAGCUGAAUUUGGCGCCCAACGUGGGGCAUAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCUUUGCCCAAACGCCCCAUCAUCAGAC--------- .((((..((((....))))......)))).....(((((((.(((((.....))))).((((((((.((....)).))))))))........)))))))........--------- ( -38.60) >DroEre_CAF1 21846 106 + 1 CGUGCAGCAGCUGAAGCUGACUUGGGCGC-CAACGUGGGGCACAUUCACCGGGAGUAAAUGGGUCUACCUAUUGGCGGGCUAUUGCCCAAGCGCCCCAUCAUCAGAG--------- .((((((((((....)))).))...))))-....(((((((.(...........((.((((((....)))))).))((((....))))..).)))))))........--------- ( -40.30) >DroYak_CAF1 19720 96 + 1 CGUGCAGCAGCUGAAGCUGAGUUUU---------GGGGAGCAGACUCAUCCGAAAUAAAUAGGUCUACCUAG--------UAUUGC---AGGACCCCAUCAUCAGACUCCAGUCUC .........((((.((((((....(---------((((((....))).(((..((((..((((....)))).--------))))..---.))).))))...)))).)).))))... ( -27.20) >consensus CGUGCAGCAGCUGAAGCUGAAUUUGGCGCCCAACGUGGGGCACAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCAUUGCCCAAACGCCCCAUCAUCAGAC_________ .((((..((((....))))......)))).....(((((((...................(((....)))......((((....))))....)))))))................. (-22.46 = -24.24 + 1.78)

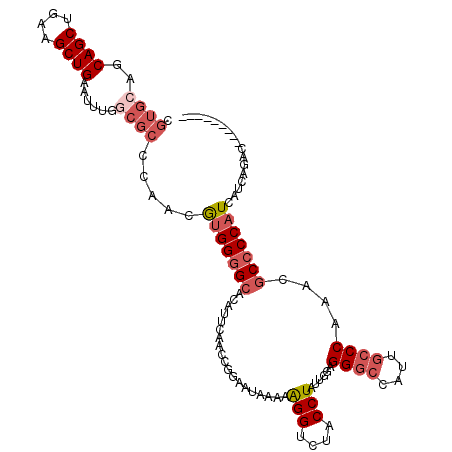

| Location | 18,031,960 – 18,032,061 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18031960 101 + 22224390 GGGGCACAUUCAACCGGAAUAAAAAGGUUCACCUAUUGGAGGGCCCUUGCCCAAACGCCCCAUCACCAGACCUGUUCGUCUCCAUCAUCAUCAGCAGUCCA (((((...(((((...((((......)))).....)))))((((....))))....))))).......(..(((((.((..........)).)))))..). ( -25.40) >DroSec_CAF1 19942 101 + 1 GGGGCAUAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCCUUGCCCAAACGCCCCAUCAUCAGACCAGCUCGUCUCCAUCAUCAUCAGCGGUCCA (((((.(((((.....)))))....(((((.((....)).)))))...........))))).......((((.(((.((..........)).))))))).. ( -29.80) >DroSim_CAF1 20271 101 + 1 GGGGCAUAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCUUUGCCCAAACGCCCCAUCAUCAGACCAGCUCGUCUCCAUCAUCAUCAGCGCUCCA (((((.(((((.....))))).((((((((.((....)).))))))))........)))))......((((......)))).................... ( -29.00) >DroEre_CAF1 21881 101 + 1 GGGGCACAUUCACCGGGAGUAAAUGGGUCUACCUAUUGGCGGGCUAUUGCCCAAGCGCCCCAUCAUCAGAGCUGCUCGGCUCCAUCAUCAUCAGCGGGCCA (((((.(...........((.((((((....)))))).))((((....))))..).))))).......(((...)))((((((..........).))))). ( -36.00) >consensus GGGGCACAUUCAACCGGAAUAAAAAGGUCUACCUAUUGGAGGGCCCUUGCCCAAACGCCCCAUCAUCAGACCAGCUCGUCUCCAUCAUCAUCAGCGGUCCA (((((..((((.....))))....(((....)))......((((....))))....))))).......(..(((((................)))))..). (-22.89 = -23.02 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:41 2006