
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,027,510 – 18,027,687 |
| Length | 177 |
| Max. P | 0.869080 |

| Location | 18,027,510 – 18,027,619 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -32.56 |
| Energy contribution | -33.76 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
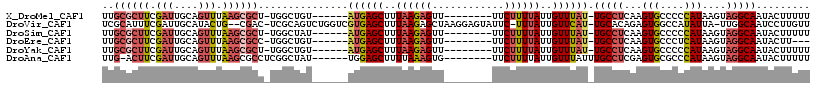
>X_DroMel_CAF1 18027510 109 + 22224390 GUGUGUGCCAAGUCGUCCUGGCGCGAGUCCUGCAAUGGAGGUUUCUUCAUUGGAGGAGGCUCCUCCAUUGGCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCUUGGCUGU ......((((((.(((...(((....)))(((((((.(((((.....((.((((((.....)))))).))((.....))))))).))))))).....)))))))))... ( -45.50) >DroSec_CAF1 16031 104 + 1 GUGUGAGCCAAGUCGUCCUGGCGCGAGUCCUGCAAUGG--GUUUCUUCAUUGG---AGGCUCCUCCAUUGGCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCUUGGCUAU .....((((((((......(((....)))..(((((((--((.....((.(((---((....))))).)))))))))))((((.(((....))).)))))))))))).. ( -44.20) >DroSim_CAF1 16347 106 + 1 GUGUGAGCCAAGUCGUCCUGGCGCGAGUCCUGCAAUGGAGGUUUCUUCAUUGG---AGGCUCCUCCAUUGGCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCUUGGCUAU .....(((((((.(((...(((....)))(((((((.(((((.....((.(((---((....))))).))((.....))))))).))))))).....)))))))))).. ( -44.50) >DroEre_CAF1 17974 106 + 1 GUGUGUGGCAAGUCGUCCUGGCGCGAGUCCUGCAAUGGAGGUUUCUUCAUUGG---AGGCUCCUCCAUUGGCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCCUGGCUGU ..............(((..(((((..(..(((((((.(((((.....((.(((---((....))))).))((.....))))))).)))))))..)..))))).)))... ( -41.30) >DroYak_CAF1 15283 106 + 1 GUGUGUACCAUGUCGUCCUGGCGCGAGUCCUGCAAUGGAGGUUUCUUCAUUGG---AGGCUCCUCCAUUGCCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCUUGGCUGU ((((......(((.(((..(((((((.....((((((((((..((((.....)---)))..))))))))))....)))))))..))).)))......))))........ ( -43.40) >consensus GUGUGUGCCAAGUCGUCCUGGCGCGAGUCCUGCAAUGGAGGUUUCUUCAUUGG___AGGCUCCUCCAUUGGCCCAUUGCGCUUCGAUUGCAGUUUAAGCGCUUGGCUGU ......(((((((.(((..(((((((......(((((((((..(((..........)))..))))))))).....)))))))..))).((.......)))))))))... (-32.56 = -33.76 + 1.20)



| Location | 18,027,510 – 18,027,619 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -27.45 |
| Energy contribution | -28.29 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18027510 109 - 22224390 ACAGCCAAGCGCUUAAACUGCAAUCGAAGCGCAAUGGGCCAAUGGAGGAGCCUCCUCCAAUGAAGAAACCUCCAUUGCAGGACUCGCGCCAGGACGACUUGGCACACAC ...(((((((((((.....((.......))(((((((..((.((((((.....)))))).)).((....))))))))).((.......))))).)).))))))...... ( -35.90) >DroSec_CAF1 16031 104 - 1 AUAGCCAAGCGCUUAAACUGCAAUCGAAGCGCAAUGGGCCAAUGGAGGAGCCU---CCAAUGAAGAAAC--CCAUUGCAGGACUCGCGCCAGGACGACUUGGCUCACAC ..((((((((((((.....((.......))((((((((.((.(((((....))---))).))......)--))))))).((.......))))).)).)))))))..... ( -35.90) >DroSim_CAF1 16347 106 - 1 AUAGCCAAGCGCUUAAACUGCAAUCGAAGCGCAAUGGGCCAAUGGAGGAGCCU---CCAAUGAAGAAACCUCCAUUGCAGGACUCGCGCCAGGACGACUUGGCUCACAC ........((((((............))))))..(((((((((((.(((((((---.(((((.((....)).))))).))).))).).))).......))))))))... ( -34.61) >DroEre_CAF1 17974 106 - 1 ACAGCCAGGCGCUUAAACUGCAAUCGAAGCGCAAUGGGCCAAUGGAGGAGCCU---CCAAUGAAGAAACCUCCAUUGCAGGACUCGCGCCAGGACGACUUGCCACACAC ...(((.(((((.....(((((((.((.((.(...).))((.(((((....))---))).))........)).))))))).....))))).)).).............. ( -33.00) >DroYak_CAF1 15283 106 - 1 ACAGCCAAGCGCUUAAACUGCAAUCGAAGCGCAAUGGGGCAAUGGAGGAGCCU---CCAAUGAAGAAACCUCCAUUGCAGGACUCGCGCCAGGACGACAUGGUACACAC ..((((..((((((............))))))......((((((((((..(.(---(....)).)...)))))))))).)).)).(..(((........)))..).... ( -32.70) >consensus ACAGCCAAGCGCUUAAACUGCAAUCGAAGCGCAAUGGGCCAAUGGAGGAGCCU___CCAAUGAAGAAACCUCCAUUGCAGGACUCGCGCCAGGACGACUUGGCACACAC ...(((..((((((............))))))....)))(((((((((....................)))))))))........(.((((((....)))))).).... (-27.45 = -28.29 + 0.84)



| Location | 18,027,585 – 18,027,687 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.67 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18027585 102 + 22224390 UUGCGCUUCGAUUGCAGUUUAAGCGCU-UGGCUGU------AUGAGCUUUAAGAGUU--------UUCUUUUAUUGUUUAU-UGCCUCAAGUGCCCCCAUAAGUAGGCAAUACUUUUU ..((((((.(((....))).)))))).-.(((...------((((((..((((((..--------..))))))..))))))-.))).....((((..(....)..))))......... ( -29.00) >DroVir_CAF1 39638 112 + 1 UCGCAUUUCGAUUGCAUACUG--CGAC-UCGCAGUCUGGUCGUGAGCUUUAAGAGCUAAGGAGUAUUC-UUUAUUGUUCAU-UGCACAGAGUGGCCAUAUUA-UUGGCAAUCCUUGUU ........(((((((.....)--))).-)))((.((((((.((((((...(((((((....)))..))-))....))))))-.)).)))).))((((.....-.)))).......... ( -30.20) >DroSim_CAF1 16419 102 + 1 UUGCGCUUCGAUUGCAGUUUAAGCGCU-UGGCUAU------AUGAGCUUUAAGAGUU--------UUCUUUUAUUGUUUAU-UGCCUCAAGUGCCCCCAUAAGUAGGCAAUACUUUUU ..((((((.(((....))).)))))).-.(((...------((((((..((((((..--------..))))))..))))))-.))).....((((..(....)..))))......... ( -29.00) >DroEre_CAF1 18046 99 + 1 UUGCGCUUCGAUUGCAGUUUAAGCGCC-UGGCUGU------AUGAGCUUUAAGAGUU--------UUCUUUUAUUGUUUAU-UGCCUCAAGUGCCCUCAUAAGUAGGCAAUACUU--- ..((((((.(((....))).)))))).-.(((...------((((((..((((((..--------..))))))..))))))-.))).....((((..(....)..))))......--- ( -28.80) >DroYak_CAF1 15355 102 + 1 UUGCGCUUCGAUUGCAGUUUAAGCGCU-UGGCUGU------AUGAGCUUUAAGAGUU--------UUCUUUUAUUGUUUAU-UGCCUCAAGUGCCCCCAUAAGUAGGCAAUACUUUUU ..((((((.(((....))).)))))).-.(((...------((((((..((((((..--------..))))))..))))))-.))).....((((..(....)..))))......... ( -29.00) >DroAna_CAF1 46270 103 + 1 UUG-ACUUCGAUUGCAGUUUAAGCGCCUCGGCUAU------UGGAGCUUUUAAAGUG--------UUCUUUUAUUGUUUAUUUGCCUCGAGUGCGCCCAUAAGUAGGCAAUACUUUUU ...-......(((((..((((.((((((((((...------..((((....((((..--------..))))....))))....)))..))).))))...))))...)))))....... ( -23.50) >consensus UUGCGCUUCGAUUGCAGUUUAAGCGCC_UGGCUGU______AUGAGCUUUAAGAGUU________UUCUUUUAUUGUUUAU_UGCCUCAAGUGCCCCCAUAAGUAGGCAAUACUUUUU ..((((((.(((....))).))))))...............((((((..((((((............))))))..)))))).(((((...(((....)))....)))))......... (-14.14 = -15.67 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:37 2006