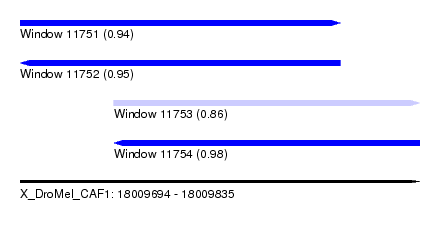
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,009,694 – 18,009,835 |
| Length | 141 |
| Max. P | 0.984174 |

| Location | 18,009,694 – 18,009,807 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.99 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009694 113 + 22224390 CG-------CAGCGGGCAGAGGCAGCGACAGAGGCGUCGACAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAAAAACACCAAAAUAGAUUCUGUAUCUG ..-------(((...((((((.(.(((((......)))).)...((((((((((.(((((((..................)))))))))))...)))))).......).))))))..))) ( -33.57) >DroSec_CAF1 21189 99 + 1 C-------------------GGCAGCGACAGAGGCGUCGGCAGCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--ACACCAAAAUAGAUUUUGUAUCUG .-------------------.((.(((((......))).)).))((((((((((.(((((((..................)))))))))).)))--)))).....(((((.....))))) ( -30.57) >DroEre_CAF1 22085 117 + 1 CGACUGAGGCAGC-GGCAGCGGCAGCGGCAGAGGCGUCGGCAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--ACACCAAAAUAGAUUCUGUAUCUG .(.((((.((..(-.((.((....)).)).)..)).)))))...((((((((((.(((((((..................)))))))))).)))--)))).....(((((.....))))) ( -39.07) >DroYak_CAF1 9038 111 + 1 CG-------CAGUGGGCAGAGGCGGCGACAGAGGCGUCGGCAUGGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--AAACCAAAAUAGAUUCUGUAUCUG ..-------(((...((((((.(((((.......))))).).((((.(((((((.(((((((..................)))))))))).)))--).))))........)))))..))) ( -37.57) >consensus CG_______CAGC_GGCAGAGGCAGCGACAGAGGCGUCGGCAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA__ACACCAAAAUAGAUUCUGUAUCUG ................((((.((((((((......)))).....((((..((((.(((((((..................))))))))))).....))))...........)))).)))) (-26.06 = -26.99 + 0.94)


| Location | 18,009,694 – 18,009,807 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009694 113 - 22224390 CAGAUACAGAAUCUAUUUUGGUGUUUUUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUGUCGACGCCUCUGUCGCUGCCUCUGCCCGCUG-------CG ..(((((((((....)))))((((((...((((((((((((...((((....)))).)))))))).))))))))))..))))((((....))))((.((........)).)-------). ( -34.80) >DroSec_CAF1 21189 99 - 1 CAGAUACAAAAUCUAUUUUGGUGU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGCUGCCGACGCCUCUGUCGCUGCC-------------------G ......(((((....)))))((((--(((.(((((((((((...((((....)))).)))))))).))))))))))((.((.((((....)))))).)).-------------------. ( -34.40) >DroEre_CAF1 22085 117 - 1 CAGAUACAGAAUCUAUUUUGGUGU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUGCCGACGCCUCUGCCGCUGCCGCUGCC-GCUGCCUCAGUCG ......(((((....)))))((((--(((.(((((((((((...((((....)))).)))))))).)))))))))).....((((......((.((.((....)).-)).))....)))) ( -37.30) >DroYak_CAF1 9038 111 - 1 CAGAUACAGAAUCUAUUUUGGUUU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACCAUGCCGACGCCUCUGUCGCCGCCUCUGCCCACUG-------CG (((...((((........((((.(--(((.(((((((((((...((((....)))).)))))))).))))))).)))).(((((((....)))))..)).))))....)))-------.. ( -33.10) >consensus CAGAUACAGAAUCUAUUUUGGUGU__UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUGCCGACGCCUCUGUCGCUGCCUCUGCC_GCUG_______CG ......(((((....)))))((((.....((((((((((((...((((....)))).)))))))).))))..))))...(((((((....)))))..))..................... (-25.38 = -25.50 + 0.12)



| Location | 18,009,727 – 18,009,835 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009727 108 + 22224390 CAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAAAAACACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUCAUCUCUGGCU------------GUAUCUGU ....((((((((((.(((((((..................)))))))))))...))))))....((((((.........((((((...........))))------------)))))))) ( -28.87) >DroSec_CAF1 21210 106 + 1 CAGCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--ACACCAAAAUAGAUUUUGUAUCUGCAGCUGCAUCAUUUGUAGCU------------GUAUCUGU ..(.((((((((((.(((((((..................)))))))))).)))--)))))...((((((.........(((((((((.....)))))))------------)))))))) ( -37.87) >DroEre_CAF1 22124 118 + 1 CAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--ACACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUCAUCUGUAGUUGUAUCUCUAUCUGUAUCUGU ....((((((((((.(((((((..................)))))))))).)))--))))....((((((...(....((((((((((.....)))))))))).)...))))))...... ( -37.27) >DroYak_CAF1 9071 100 + 1 CAUGGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA--AAACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUCAUCU------------------GUAUCUGU ..((((.(((((((.(((((((..................)))))))))).)))--).))))....((((.....))))((((.((((.....)------------------))).)))) ( -27.97) >consensus CAUCGUGUUUAUCUAGGCAGCCCAACUAUUUAUAAUAAAAGGCUGCCAGAUUAA__ACACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUCAUCUGUAGCU____________GUAUCUGU ....((((..((((.(((((((..................))))))))))).....))))....((((((..((((.(....).))))..))))))........................ (-21.83 = -22.64 + 0.81)


| Location | 18,009,727 – 18,009,835 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.84 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009727 108 - 22224390 ACAGAUAC------------AGCCAGAGAUGAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUGUUUUUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUG ........------------.(((((((..(((....(((......))).)))..)))))))((((...((((((((((((...((((....)))).)))))))).))))))))...... ( -30.70) >DroSec_CAF1 21210 106 - 1 ACAGAUAC------------AGCUACAAAUGAUGCAGCUGCAGAUACAAAAUCUAUUUUGGUGU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGCUG .((((..(------------((((.((.....)).))))).((((.....))))..))))((((--(((.(((((((((((...((((....)))).)))))))).)))))))))).... ( -35.20) >DroEre_CAF1 22124 118 - 1 ACAGAUACAGAUAGAGAUACAACUACAGAUGAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUGU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUG .......((((...((((.((.(....).)).(((....)))........))))..))))((((--(((.(((((((((((...((((....)))).)))))))).)))))))))).... ( -32.80) >DroYak_CAF1 9071 100 - 1 ACAGAUAC------------------AGAUGAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUUU--UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACCAUG ..((((..------------------......(((....)))........))))....((((.(--(((.(((((((((((...((((....)))).)))))))).))))))).)))).. ( -28.89) >consensus ACAGAUAC____________AGCUACAGAUGAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUGU__UUAAUCUGGCAGCCUUUUAUUAUAAAUAGUUGGGCUGCCUAGAUAAACACGAUG ................................(((....)))....(((((....)))))((((.....((((((((((((...((((....)))).)))))))).))))..)))).... (-24.84 = -24.90 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:28 2006