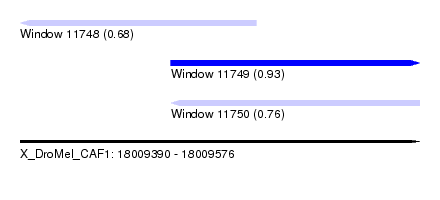
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,009,390 – 18,009,576 |
| Length | 186 |
| Max. P | 0.934013 |

| Location | 18,009,390 – 18,009,500 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -13.96 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009390 110 - 22224390 AGAUUAAGUGCCUGGCUGGGAAAUAUUUCCAGCUCAUUUGGGCAAGAAAAUCGGACUGCUUUCCCUGAGAAUCUAAACAGCC-AAAACUAAGGAAAUUCAUUUAAAUUA---------AU ......(((...((((((((((....)))).((((....)))).(((...((((..........))))...)))...)))))-)..)))....................---------.. ( -23.70) >DroSec_CAF1 20884 110 - 1 AGAUUAAGUGCCUGGCUGGAAAAUAUUUCCAGCUCAUUUGGGCAAGAAAAUCGCACUGCUUUCCUUGGAAAUCUUAGCAGCU-AAAACCAAGGCAAUUAAUUUAAGUUA---------AU (((((((.(((((((((((((.....))))))))....(((((.........)).((((((((....))).....)))))..-....)))))))).)))))))......---------.. ( -31.70) >DroEre_CAF1 21773 116 - 1 AAAUUAAAUGCCUGGCUGGGAAAAUUUUCCAGUUGAUUUGGGCAAGAAAAACCGAC----UUCCUUGGGAAUCUCAGCAGCUUGAAAGCAAGCCGUUUAAUUUACAUUUUCAACUUUUAG ((((((((((....(((((((....(((((((..((.((((..........)))).----))..)))))))))))))).(((((....)))))))))))))))................. ( -30.90) >DroYak_CAF1 8746 100 - 1 AAACUCACAGCUGGGCUGGGAAAUAAUUUCAGUUGAUUUGGGCAAGAAAAUC-----------UGUGGGAAUCUUAGCAGUUUAAAGUCAAGCCAUUUAAUUUAGCUUU---------AU ...((((((....(((((..(.....)..)))))(((((........)))))-----------)))))).............(((((..(((........)))..))))---------). ( -18.20) >consensus AAAUUAAGUGCCUGGCUGGGAAAUAUUUCCAGCUCAUUUGGGCAAGAAAAUCGGAC____UUCCUUGGGAAUCUUAGCAGCU_AAAACCAAGCCAAUUAAUUUAAAUUA_________AU .........((((((((((((.....)))))))).....))))............................................................................. (-13.96 = -13.40 + -0.56)


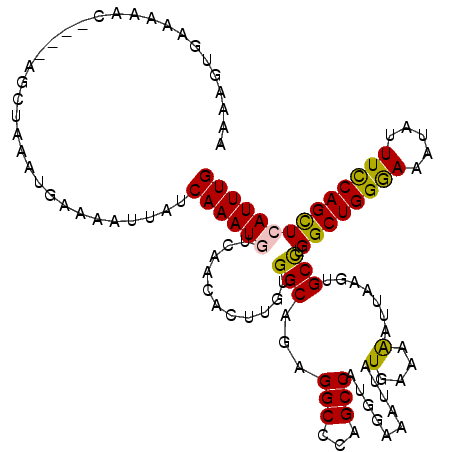
| Location | 18,009,460 – 18,009,576 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -17.62 |
| Energy contribution | -19.75 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009460 116 + 22224390 CAAAUGAGCUGGAAAUAUUUCCCAGCCAGGCACUUAAUCUUUCAUAAUUUCCAUGGCUGGGCCUUUGCCACAAGUGUUGACAUUUGAUAAUUUUCAUUUAGCC----GUUUUUCACUUUU .(((((.((((((.......((((((((((...(((........)))...)).)))))))).........((((((....))))))..........)))))))----))))......... ( -29.20) >DroSec_CAF1 20954 116 + 1 CAAAUGAGCUGGAAAUAUUUUCCAGCCAGGCACUUAAUCUUUCAUAAUUUCCAUGGCUGUGCCUCUGCCGCAAGUGUUGACAUUUGAUAAUUUUCAUUUAGGC----GUUUUUCACUUUU ....((.(((((((.....)))))))))(((((.........................)))))...(((.((((((....)))))).(((.......))))))----............. ( -26.41) >DroEre_CAF1 21849 120 + 1 CAAAUCAACUGGAAAAUUUUCCCAGCCAGGCAUUUAAUUUUUCAUAAAUUGCAUGGCUGGGCCGCAGCCACAAGUGUUGACAUUUGAUAAUUUUCAUUUAGAUUUAUUUUUUUUGCUUUU .(((((.....(((((((..((((((((.(((((((........)))).))).)))))))).........((((((....))))))..))))))).....)))))............... ( -31.30) >DroYak_CAF1 8806 116 + 1 CAAAUCAACUGAAAUUAUUUCCCAGCCCAGCUGUGAGUUUUUCAUAAUUUUCAUGGCUUGGCCAGUGCCACAAGUGUUGUCAUUUGAUCAUUUUCAUUUAGCU----GCUUUUCACUUUU ...(((((.(((............(((.(((((((((..((....))..))))))))).)))(((..(.....)..)))))).)))))...............----............. ( -23.50) >consensus CAAAUCAACUGGAAAUAUUUCCCAGCCAGGCACUUAAUCUUUCAUAAUUUCCAUGGCUGGGCCUCUGCCACAAGUGUUGACAUUUGAUAAUUUUCAUUUAGCC____GUUUUUCACUUUU .........(((((((....((((((((((.(((((........))))).)).)))))))).........((((((....))))))...)))))))........................ (-17.62 = -19.75 + 2.12)



| Location | 18,009,460 – 18,009,576 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -16.26 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18009460 116 - 22224390 AAAAGUGAAAAAC----GGCUAAAUGAAAAUUAUCAAAUGUCAACACUUGUGGCAAAGGCCCAGCCAUGGAAAUUAUGAAAGAUUAAGUGCCUGGCUGGGAAAUAUUUCCAGCUCAUUUG ..((((((.....----(((....(((......)))...)))........(((.(((..((((((((.((...(((........)))...)))))))))).....))))))..)))))). ( -28.90) >DroSec_CAF1 20954 116 - 1 AAAAGUGAAAAAC----GCCUAAAUGAAAAUUAUCAAAUGUCAACACUUGCGGCAGAGGCACAGCCAUGGAAAUUAUGAAAGAUUAAGUGCCUGGCUGGAAAAUAUUUCCAGCUCAUUUG ..((((((.....----(((..(((....)))..(((.((....)).))).)))..((((((..(....).((((......))))..)))))).(((((((.....))))))))))))). ( -27.50) >DroEre_CAF1 21849 120 - 1 AAAAGCAAAAAAAAUAAAUCUAAAUGAAAAUUAUCAAAUGUCAACACUUGUGGCUGCGGCCCAGCCAUGCAAUUUAUGAAAAAUUAAAUGCCUGGCUGGGAAAAUUUUCCAGUUGAUUUG ..............((((((.....(((((((.......((((.(....))))).....((((((((.(((.((((........))))))).))))))))..))))))).....)))))) ( -30.40) >DroYak_CAF1 8806 116 - 1 AAAAGUGAAAAGC----AGCUAAAUGAAAAUGAUCAAAUGACAACACUUGUGGCACUGGCCAAGCCAUGAAAAUUAUGAAAAACUCACAGCUGGGCUGGGAAAUAAUUUCAGUUGAUUUG ..((.(((((...----.((((........((.((....)))).......)))).(..(((.(((..(((..............)))..))).)))..).......))))).))...... ( -21.40) >consensus AAAAGUGAAAAAC____AGCUAAAUGAAAAUUAUCAAAUGUCAACACUUGUGGCAGAGGCCCAGCCAUGGAAAUUAUGAAAAAUUAAGUGCCUGGCUGGGAAAUAUUUCCAGCUCAUUUG ..................................((((((...........(((...(((...)))..........(....).......))).((((((((.....)))))))))))))) (-16.26 = -15.70 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:25 2006