| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,004,320 – 18,004,412 |
| Length | 92 |
| Max. P | 0.992209 |

| Location | 18,004,320 – 18,004,412 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -13.87 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
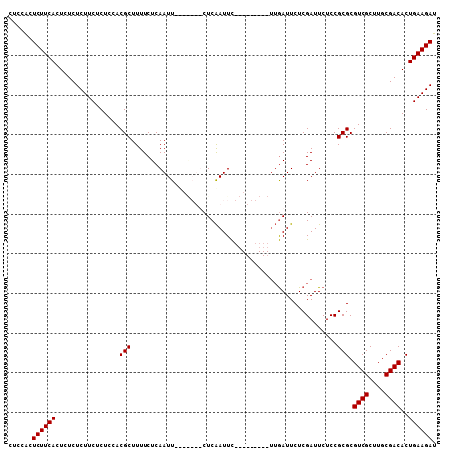
>X_DroMel_CAF1 18004320 92 + 22224390 CUCCACUCUUCACUUUCUCUUCUCUCCACGCUUUUCUCAAUU-------CUCAAUUC---------UUGAUUCUCGAUUCUCCGCGCGUCGCUUGCGACACUGAAGAU ......((((((................(((.....((....-------.(((....---------.))).....))......))).((((....))))..)))))). ( -12.70) >DroSec_CAF1 16036 99 + 1 CUCCGCUCUUCACUCUCUCUUCUCUCCACGCUUUUCUCAAUUCUCAAUUCUCAAUUC---------UUGAUUCUCGAUUCUCCGCGCGUCGCUUGCGACACUGAAGAU ......((((((................(((.....((.....((((..........---------)))).....))......))).((((....))))..)))))). ( -12.70) >DroSim_CAF1 4943 92 + 1 CUCCACUCUUCACUCUCUCUUCUCUCCACGCUUUUCUCAAUU-------CUCAAUUC---------UUGAUUCUCGAUUCUCCGCGCGUCGCUUGCGACACUGAAGAU ......((((((................(((.....((....-------.(((....---------.))).....))......))).((((....))))..)))))). ( -12.70) >DroEre_CAF1 16849 106 + 1 CUCCACUCUUCACUUUCUCUUCUCUCCACGCUUUUCUCAAUUCCCGAUUCUCGAUUUUUGAUUCUC--AAUUCUCGAUUCUCCGCGCGUCGCUUGCGACACUGAAGAU ......((((((................(((.....((.......))...((((...(((.....)--))...))))......))).((((....))))..)))))). ( -17.40) >consensus CUCCACUCUUCACUCUCUCUUCUCUCCACGCUUUUCUCAAUU_______CUCAAUUC_________UUGAUUCUCGAUUCUCCGCGCGUCGCUUGCGACACUGAAGAU ......((((((................(((....................................................))).((((....))))..)))))). (-11.22 = -11.22 + -0.00)


| Location | 18,004,320 – 18,004,412 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -19.96 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
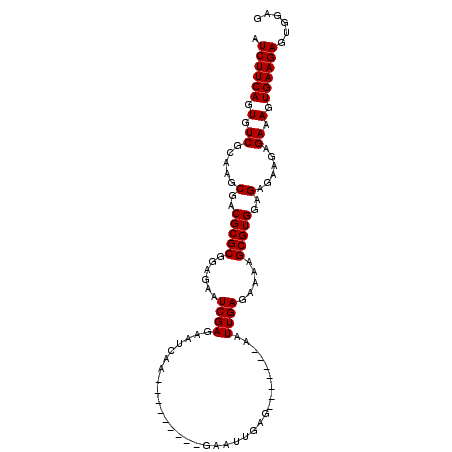
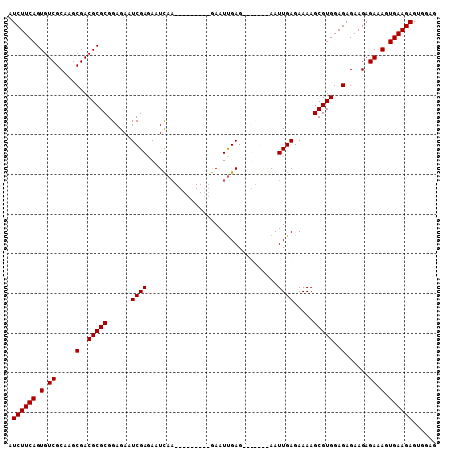
>X_DroMel_CAF1 18004320 92 - 22224390 AUCUUCAGUGUCGCAAGCGACGCGCGGAGAAUCGAGAAUCAA---------GAAUUGAG-------AAUUGAGAAAAGCGUGGAGAGAAGAGAAAGUGAAGAGUGGAG .((((((.(.((.....(..(((((......((((...(((.---------....))).-------..)))).....)))))..)......)).).))))))...... ( -23.60) >DroSec_CAF1 16036 99 - 1 AUCUUCAGUGUCGCAAGCGACGCGCGGAGAAUCGAGAAUCAA---------GAAUUGAGAAUUGAGAAUUGAGAAAAGCGUGGAGAGAAGAGAGAGUGAAGAGCGGAG .((((((.(.((.....(..(((((......((((...((((---------..........))))...)))).....)))))..)......)).).))))))...... ( -26.50) >DroSim_CAF1 4943 92 - 1 AUCUUCAGUGUCGCAAGCGACGCGCGGAGAAUCGAGAAUCAA---------GAAUUGAG-------AAUUGAGAAAAGCGUGGAGAGAAGAGAGAGUGAAGAGUGGAG .((((((.(.((.....(..(((((......((((...(((.---------....))).-------..)))).....)))))..)......)).).))))))...... ( -25.70) >DroEre_CAF1 16849 106 - 1 AUCUUCAGUGUCGCAAGCGACGCGCGGAGAAUCGAGAAUU--GAGAAUCAAAAAUCGAGAAUCGGGAAUUGAGAAAAGCGUGGAGAGAAGAGAAAGUGAAGAGUGGAG .((((((.(.((.....(..(((((......((((...((--((...((.......))...))))...)))).....)))))..)......)).).))))))...... ( -25.50) >consensus AUCUUCAGUGUCGCAAGCGACGCGCGGAGAAUCGAGAAUCAA_________GAAUUGAG_______AAUUGAGAAAAGCGUGGAGAGAAGAGAAAGUGAAGAGUGGAG .((((((.(.((.....(..(((((......((((.................................)))).....)))))..)......)).).))))))...... (-19.96 = -19.96 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:18 2006