| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,000,622 – 18,000,741 |
| Length | 119 |
| Max. P | 0.837869 |

| Location | 18,000,622 – 18,000,741 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -22.73 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
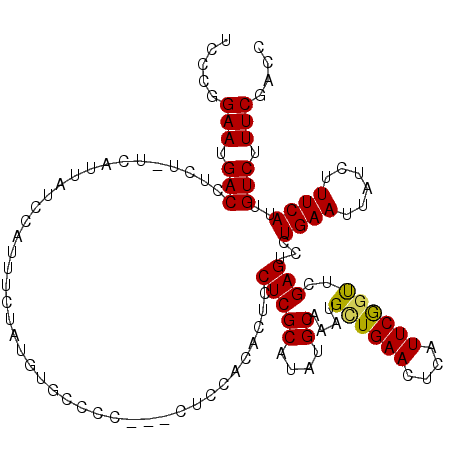
>X_DroMel_CAF1 18000622 119 + 22224390 GGUCGAAAGACAAUGAAAGAUAAUUCAGGACUCGAACCGAAUGAGUUCAGCAUUUGCAUAUGCGAGAAGUGUGGAGUGUGGGGCACAUAGAAAUGGAUAAUGA-AGAGGUCAUUCUGGGA .(((....)))..((((......)))).........((((((((.(((.((((((((....))))...))))...(((.....))).................-.))).)))))).)).. ( -27.50) >DroSec_CAF1 12550 116 + 1 GGUCGAAAGACAAUGAAAGAUAAUUCAGGACUCGAACCGAAUGAGUUCACCAUUUGCAUAUGCGAGGAGUGUGGAG---GGGUCACAUAGAAAUGGAUAAUGA-AGAGGUCAUUCCGGGA .(((....)))...(((.(((..(((((((((((.......))))))).((((((((....)).....((((((..---...))))))..))))))....)))-)...))).)))..... ( -31.00) >DroEre_CAF1 13369 116 + 1 GGUCGAAAGACAAUGAAAGAUAAUUCAGAACUCGAGCCGAAUGAGUUCAACAUUUGCAUAUGCGAGGAGUGUGGAG---GGGGCACAUAGAAAUGGAUAAUGA-AGAGGUCAUUCCCGGA .(((....)))...(((.(((..(((((((((((.......)))))))..(((((((....)).....(((((...---....)))))..))))).....)))-)...))).)))..... ( -27.70) >DroYak_CAF1 158 96 + 1 GGUCGAAAGACAAUGAAAGAUAAUUCAGGACUCGCACUGAAUGAGUUCAGCAUUUGCAUAUGCGAGG------------------------AAUGGAUAAUGGCGGAGGUCAUUCCGGGA .(((....)))...........(((((...((((((.((((....))))((....))...)))))).------------------------..))))).....(((((....)))))... ( -27.80) >consensus GGUCGAAAGACAAUGAAAGAUAAUUCAGGACUCGAACCGAAUGAGUUCAGCAUUUGCAUAUGCGAGGAGUGUGGAG___GGGGCACAUAGAAAUGGAUAAUGA_AGAGGUCAUUCCGGGA .(((....)))..((((......))))...((((....((((((.(((....(((((....)))))..(((((..........))))).................))).)))))))))). (-22.73 = -23.10 + 0.37)


| Location | 18,000,622 – 18,000,741 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.60 |
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18000622 119 - 22224390 UCCCAGAAUGACCUCU-UCAUUAUCCAUUUCUAUGUGCCCCACACUCCACACUUCUCGCAUAUGCAAAUGCUGAACUCAUUCGGUUCGAGUCCUGAAUUAUCUUUCAUUGUCUUUCGACC .....((((((....(-(((.....(((((.(((((((...................))))))).))))).)))).))))))(((.((((...((((......))))......))))))) ( -20.11) >DroSec_CAF1 12550 116 - 1 UCCCGGAAUGACCUCU-UCAUUAUCCAUUUCUAUGUGACCC---CUCCACACUCCUCGCAUAUGCAAAUGGUGAACUCAUUCGGUUCGAGUCCUGAAUUAUCUUUCAUUGUCUUUCGACC ...(((((.((((((.-.......((((((((((((((...---...........))))))).).)))))).(((((.....))))))))...((((......))))..))))))))... ( -24.04) >DroEre_CAF1 13369 116 - 1 UCCGGGAAUGACCUCU-UCAUUAUCCAUUUCUAUGUGCCCC---CUCCACACUCCUCGCAUAUGCAAAUGUUGAACUCAUUCGGCUCGAGUUCUGAAUUAUCUUUCAUUGUCUUUCGACC .....((((((....(-(((.....(((((.(((((((...---.............))))))).))))).)))).))))))((.(((((...((((......))))......))))))) ( -20.79) >DroYak_CAF1 158 96 - 1 UCCCGGAAUGACCUCCGCCAUUAUCCAUU------------------------CCUCGCAUAUGCAAAUGCUGAACUCAUUCAGUGCGAGUCCUGAAUUAUCUUUCAUUGUCUUUCGACC ...(((((.(((.................------------------------.(((((((..((....))((((....)))))))))))...((((......))))..))))))))... ( -19.60) >consensus UCCCGGAAUGACCUCU_UCAUUAUCCAUUUCUAUGUGCCCC___CUCCACACUCCUCGCAUAUGCAAAUGCUGAACUCAUUCGGUUCGAGUCCUGAAUUAUCUUUCAUUGUCUUUCGACC .....(((.(((..........................................(((((....))....((((((....))))))..)))...((((......))))..))).))).... (-13.38 = -13.12 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:15 2006