
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,000,345 – 18,000,504 |
| Length | 159 |
| Max. P | 0.984386 |

| Location | 18,000,345 – 18,000,464 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -28.41 |
| Energy contribution | -27.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18000345 119 + 22224390 AUGCGAGGCGGGGGGGCGUGGCCCUCCCAAAGGGG-GUGAGCGGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAAAAAGAAAGC .(((.(((((...((((((.((((((((...))))-).).)).))))))...))))).)))...((((((...((((.......)))).))))))......................... ( -43.00) >DroSec_CAF1 12292 95 + 1 -----------------------CUCCCAAAGGGG-CUGAGCGGCGUUUAAACGUUUGUUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAA-AAGAAAGC -----------------------((((((.....(-((.((((((((....))).)))))....((((((...((((.......)))).)))))).)))....))))))..-........ ( -32.00) >DroEre_CAF1 13098 117 + 1 AUGCGGAGCAGG-GUGCGUGGUCCUCCCAAAGGGGAGGGAGCAGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCGAUGGCGAUGGCCAAGCAAAAUGGGGGAAAA--AAAGC .(((.((((..(-.(((....(((((((....))))))).))).)((....)))))).)))...(((((((..((((.......)))))))))))..................--..... ( -40.30) >consensus AUGCG__GC_GG_G_GCGUGG_CCUCCCAAAGGGG_GUGAGCGGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAAAAAGAAAGC .......................((((((.............(((((....)))))........(((((((..((((.......)))))))))))........))))))........... (-28.41 = -27.97 + -0.44)

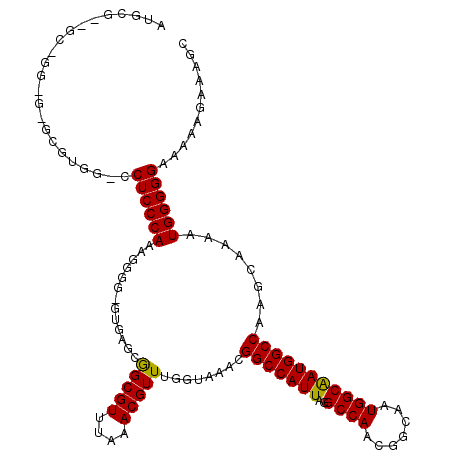
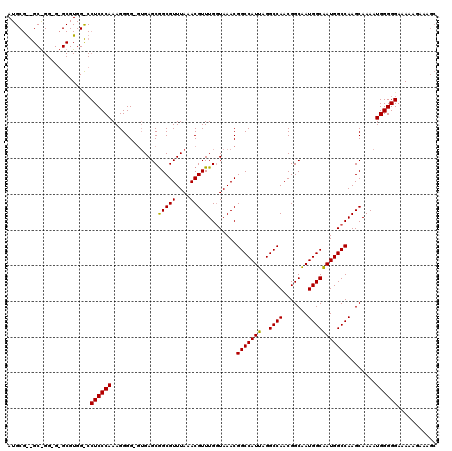
| Location | 18,000,345 – 18,000,464 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18000345 119 - 22224390 GCUUUCUUUUUCCCCCAUUUUGCUUGGCCAUUGCCAUUGCCGUUGGCCUAAUGGCCGUUUACCAAACGUUUAAACGCCGCUCAC-CCCCUUUGGGAGGGCCACGCCCCCCCGCCUCGCAU ((...................((..(((((((((((.......))))..)))))))(((((.........)))))...))....-.......(((.((((...)))).))).....)).. ( -33.40) >DroSec_CAF1 12292 95 - 1 GCUUUCUU-UUCCCCCAUUUUGCUUGGCCAUUGCCAUUGCCGUUGGCCUAAUGGCCGUUUAACAAACGUUUAAACGCCGCUCAG-CCCCUUUGGGAG----------------------- ........-....((((....(((.(((((((((((.......))))..)))))))((((((.......)))))).......))-).....))))..----------------------- ( -27.00) >DroEre_CAF1 13098 117 - 1 GCUUU--UUUUCCCCCAUUUUGCUUGGCCAUCGCCAUCGCCGUUGGCCUAAUGGCCGUUUACCAAACGUUUAAACGCUGCUCCCUCCCCUUUGGGAGGACCACGCAC-CCUGCUCCGCAU .....--..............((..((((((.((((.......))))...))))))(((((.........)))))))(((..((((((....)))))).....((..-...))...))). ( -32.80) >consensus GCUUUCUUUUUCCCCCAUUUUGCUUGGCCAUUGCCAUUGCCGUUGGCCUAAUGGCCGUUUACCAAACGUUUAAACGCCGCUCAC_CCCCUUUGGGAGG_CCACGC_C_CC_GC__CGCAU ..........(((((((........(((((((((((.......))))..)))))))(((((.........)))))................)))).)))..................... (-23.81 = -24.70 + 0.89)


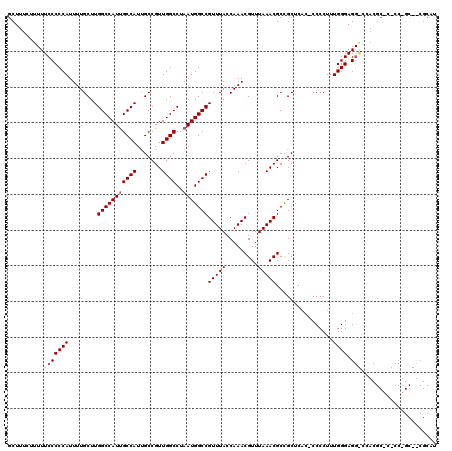
| Location | 18,000,384 – 18,000,504 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -25.41 |
| Energy contribution | -24.74 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18000384 120 + 22224390 GCGGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAAAAAGAAAGCAAACAGUGAACAUUUUAUGCCAGCAAAACAAAGAACAUUU (((((((......(((((((.....(((((...(((...))).)))))....)))))))((((((.........................))))))))))).))................ ( -30.81) >DroSec_CAF1 12308 119 + 1 GCGGCGUUUAAACGUUUGUUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAA-AAGAAAGCAAACAGAGAACAUUUUAUGCCAGAAAAACAAAGAACAUUU ..(((((..((..(((((((....((((....))))....((..((((....)))).))............-.....))))))).(....).))..)))))................... ( -26.20) >DroEre_CAF1 13137 118 + 1 GCAGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCGAUGGCGAUGGCCAAGCAAAAUGGGGGAAAA--AAAGCAAACAGAGAACAUUUUAUGCCAGCAAAACAAAGAACAUUU (((((((....)))))(((((...(((((((..((((.......)))))))))))....((((((........--...............)))))).)))))))................ ( -27.90) >consensus GCGGCGUUUAAACGUUUGGUAAACGGCCAUUAGGCCAACGGCAAUGGCAAUGGCCAAGCAAAAUGGGGGAAAAAGAAAGCAAACAGAGAACAUUUUAUGCCAGCAAAACAAAGAACAUUU .....((((....((((.((....((((....))))...((((.((((....))))...((((((.........................)))))).)))).)).))))...)))).... (-25.41 = -24.74 + -0.66)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:14 2006