
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,993,990 – 17,994,209 |
| Length | 219 |
| Max. P | 0.799561 |

| Location | 17,993,990 – 17,994,102 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.73 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.04 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
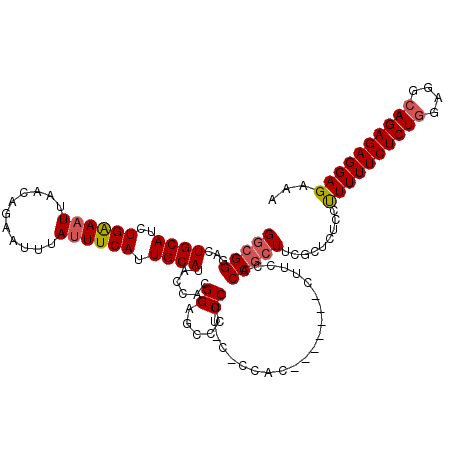
>X_DroMel_CAF1 17993990 112 + 22224390 GACGGGACUGCAUCUGAAAUUAACAGAAUUUAUUUCAUUGCAUACCACGGAGCCCCCUU-CGCCAC-------CUUCCACCGCUUCGCUCUCCUUUUUCUCUGGUGGCAGAGAGGAGAAA ...((...((((..((((((...........)))))).))))..)).((((((......-......-------........)))))).(((((....((((((....))))))))))).. ( -28.05) >DroSec_CAF1 6069 120 + 1 GGCGGGAUUGCAUCUGAAAUUAACAGAAUUUAAUUCAUUGCAUACUACGGAGCCCCCUUUCCCCACCUUACCCCUUCCACCGCUUCGCUCUCCUUUUUCUCUGGAGGCAGAGAGGAGAAA (((((...((((..((((.((((......)))))))).))))......((((......)))).................)))))....(((((....((((((....))))))))))).. ( -28.90) >DroEre_CAF1 7059 89 + 1 GGCGGGACUGCAUCUGGAAUUAACAGAAUUUAUUUCAUUGCAUAGCA-------------------------------ACCCCUUCCCGCCUCCUUUUCUCUGGCGGAAGAGAGGAGGUA (((((((.....((((.......))))..........((((...)))-------------------------------).....))))))).((((((((((......)))))))))).. ( -33.10) >consensus GGCGGGACUGCAUCUGAAAUUAACAGAAUUUAUUUCAUUGCAUACCACGGAGCCCCCUU_C_CCAC_______CUUCCACCGCUUCGCUCUCCUUUUUCUCUGGAGGCAGAGAGGAGAAA (((((...((((..((((((...........)))))).))))......((.....))......................))))).........((((((((((....))))))))))... (-17.04 = -18.10 + 1.06)

| Location | 17,994,030 – 17,994,142 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17994030 112 + 22224390 CAUACCACGGAGCCCCCUU-CGCCAC-------CUUCCACCGCUUCGCUCUCCUUUUUCUCUGGUGGCAGAGAGGAGAAAAAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAAC .......(((((....)))-))....-------.......(((.....(((((....((((((....)))))))))))...........((((...........))))....)))..... ( -24.70) >DroSec_CAF1 6109 120 + 1 CAUACUACGGAGCCCCCUUUCCCCACCUUACCCCUUCCACCGCUUCGCUCUCCUUUUUCUCUGGAGGCAGAGAGGAGAAAAAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAAC ........(((((............................)))))(((((((....((((((....)))))))))))...........((((...........))))....))...... ( -23.69) >DroEre_CAF1 7099 89 + 1 CAUAGCA-------------------------------ACCCCUUCCCGCCUCCUUUUCUCUGGCGGAAGAGAGGAGGUAAAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAAC ....((.-------------------------------..........(((((((((..((.....))..)))))))))..........((((...........))))....))...... ( -24.00) >consensus CAUACCACGGAGCCCCCUU_C_CCAC_______CUUCCACCGCUUCGCUCUCCUUUUUCUCUGGAGGCAGAGAGGAGAAAAAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAAC ........................................(((..........((((((((((....))))))))))............((((...........))))....)))..... (-15.65 = -16.10 + 0.45)



| Location | 17,994,102 – 17,994,209 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
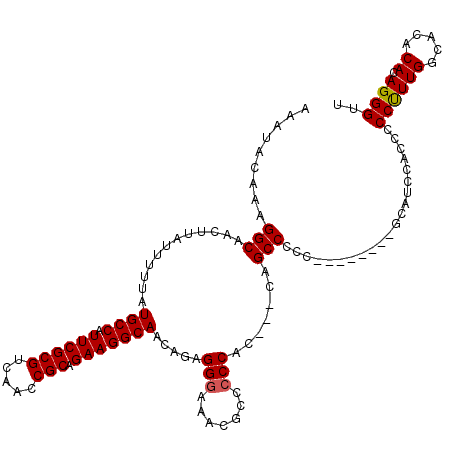
>X_DroMel_CAF1 17994102 107 + 22224390 AAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAACCGCAGAAGGCAACAGAGGAAAACGCCCCCAC---CAGCC--C--------GCAUCCACCCCCUUUGGCACACACAGGGUU .........(((............((((.((((((.....))).)))))))...(.((........)))..---..)))--.--------.........((((.((.....)).)))).. ( -23.20) >DroSec_CAF1 6189 111 + 1 AAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAACCGCAGAAGGCAACAGAGGGAACCGCCCCCACAUCCAGCCCCC--------ACAUCCACC-CCUUUGGCACACACAGGGUU .........(((............((((.((((((.....))).))))))).....(((.......))).......)))...--------........(-(((.((.....)).)))).. ( -26.70) >DroEre_CAF1 7148 116 + 1 AAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAACCGCAGAAGGCAACACAGGGAAACGCCCCCAC----AGCCCCCAGUAUCCAGUAUCCAACCCCCUUGGCACACACAGGGUU ..((((...(((............((((.((((((.....))).))))))).....(((.......)))..----.)))....)))).............((((((.....)).)))).. ( -26.30) >consensus AAAUACAAAGGCAACUUAUUUUUAUGCCAUUCGCGUCAACCGCAGAAGGCAACAGAGGGAAACGCCCCCAC___CAGCCCCC________GCAUCCACCCCCUUUGGCACACACAGGGUU .........(((............((((.((((((.....))).))))))).....(((.......))).......))).....................((((((.....)).)))).. (-21.62 = -21.73 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:08 2006