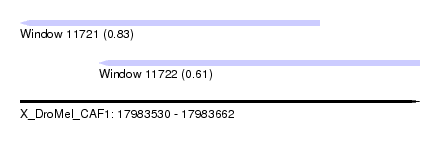
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,983,530 – 17,983,662 |
| Length | 132 |
| Max. P | 0.829883 |

| Location | 17,983,530 – 17,983,629 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -30.21 |
| Energy contribution | -31.27 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17983530 99 - 22224390 -------GCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCAAACACCUCCGUGGAUCCCCCGGUUGCUCAAUAUCUCU-------------CUAGCUUCGGUU-UUUCUCCCAUUUUC -------((((((((((((((((.(((((.....))))).)))..(((....)))........)))))).........))-------------)))))...((..-......))...... ( -29.80) >DroSim_CAF1 14177 118 - 1 CUCCCCAGCUGGAGGCAACCUGGUCGGGAGCGAAUUCCGGCCAGACACCUCCGUGGAUCCCCCGGUUGCUCAAGCUCUCAGUUUCGCUGCG-AGUAGCUUCGGUU-UUUUUCCCAUUUCC ..........(((((....((((((((((.....))))))))))...)))))(.(((....((((..(((...((((.(((.....))).)-)))))).))))..-....))))...... ( -43.60) >DroEre_CAF1 15224 118 - 1 UUCCCCAGCUGGAGGCAACCUGGUCGGGAGCGGAUUCCGACCAGACACCUCCGUGGAUCCCCCGGUUGCUCCAUCUCCCGGUUUCUCUGGA-CGUAGCUCCGGCU-UUUCUCCCAUUUCC ......(((((((((((((((((((((((.....)))))))))).......((.((....)))))))))........((((.....)))).-.....))))))))-.............. ( -43.90) >DroYak_CAF1 13534 120 - 1 CUGCCCAGCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCGGACACCUUCGUGGAUCCCCCGGCUGUUCAAUCUCUCAGUUUCUCUGGGUAGUAGCUUCGGUUUUUUCUCCCAUUUUC .((..(((((((.((....((((.(((((.....))))).)))).(((....)))....)))))))))..))...............((((.((.(((....)))....))))))..... ( -38.30) >consensus CUCCCCAGCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCAGACACCUCCGUGGAUCCCCCGGUUGCUCAAUCUCUCAGUUUCUCUGGG_AGUAGCUUCGGUU_UUUCUCCCAUUUCC ......(((((((((((((((((((((((.....)))))))))..(((....)))........))))))........((((.....)))).......))))))))............... (-30.21 = -31.27 + 1.06)

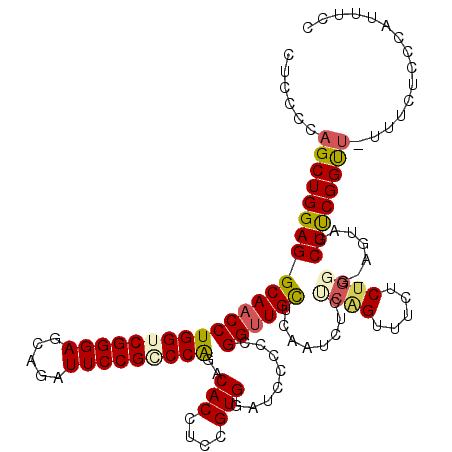

| Location | 17,983,556 – 17,983,662 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17983556 106 - 22224390 CCAGCUCCCAGUUUCUUGGC-UCCGCCAUUUUUU-------------GCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCAAACACCUCCGUGGAUCCCCCGGUUGCUCAAUAUCUCU ....((((.(((....((((-...))))......-------------)))))))(((((((((.(((((.....))))).)))..(((....)))........))))))........... ( -32.60) >DroSim_CAF1 14215 120 - 1 CCAGCCCGCAGUUCCUUGGGUUCCGCACUUUUUUUGGUGUCUCCCCAGCUGGAGGCAACCUGGUCGGGAGCGAAUUCCGGCCAGACACCUCCGUGGAUCCCCCGGUUGCUCAAGCUCUCA ..(((..((((..((..(((.(((((.......((((.......))))..(((((....((((((((((.....))))))))))...)))))))))).)))..))))))....))).... ( -46.60) >DroEre_CAF1 15262 117 - 1 CCAGCCCGCAGUUAUUUGGU-UCCGCACUU-UU-UAGUGUUUCCCCAGCUGGAGGCAACCUGGUCGGGAGCGGAUUCCGACCAGACACCUCCGUGGAUCCCCCGGUUGCUCCAUCUCCCG .....((((......((((.-...(((((.-..-.)))))....))))..(((((....((((((((((.....))))))))))...))))))))).......((.....))........ ( -40.70) >DroYak_CAF1 13574 118 - 1 CCAGCCGGCAGUCCUUUGAU-UCCGCACUUUUU-UAGUGUCUGCCCAGCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCGGACACCUUCGUGGAUCCCCCGGCUGUUCAAUCUCUCA .(((((((..((((..(((.-((((((((....-.))))(((((((.((..(.(....))..))..)).))))).......)))).....))).))))...)))))))............ ( -41.50) >consensus CCAGCCCGCAGUUCCUUGGU_UCCGCACUUUUU_UAGUGUCUCCCCAGCUGGAGGCAACCUGGUCGGGAGCAGAUUCCGCCCAGACACCUCCGUGGAUCCCCCGGUUGCUCAAUCUCUCA .(((((..(((....)))...((((((((......)))............(((((....((((((((((.....))))))))))...))))))))))......)))))............ (-32.39 = -32.95 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:01 2006