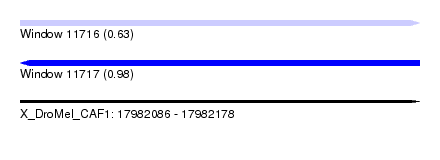
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,982,086 – 17,982,178 |
| Length | 92 |
| Max. P | 0.984450 |

| Location | 17,982,086 – 17,982,178 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
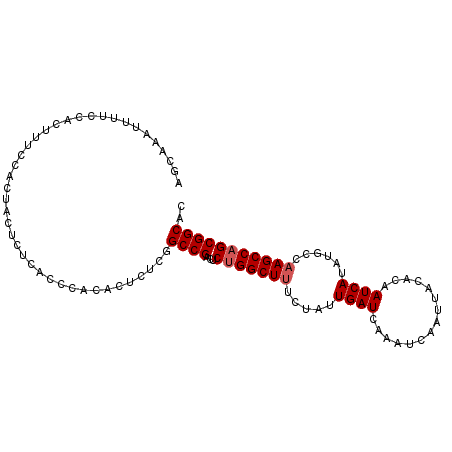
>X_DroMel_CAF1 17982086 92 + 22224390 GUGCCGCUGGCUUGGCAUAUGAUUGUGUAAUUGAUUUGAUCAAUAGAAAGCCAGAGUCGGCCGAGAGUGCGGGUG----------AAAGUGGAAAAUUUGCU ...((((((((((.(((((....))))).((((((...))))))...)))))....(((.(((......))).))----------).))))).......... ( -25.70) >DroSec_CAF1 12994 100 + 1 GUGCCGCUGGCUUGGCAUAUGAUUGUGUAAUUGAUUUGAUCA-UAGA-AGCCAGAGUCGGCCGAGAGAGUGGGUGAGAGUAGUGGAAAGUGGAAAAUUUGCU ..((((((((((((((.((((((((.((.....)).))))))-))..-.)))).)))))))(....)....)))......((..((...(....).))..)) ( -26.00) >DroSim_CAF1 12713 102 + 1 GUGCCGCUGGCUUGGCAUAUGAUUGUGUAAUUGAUUUGAUCAAUAGAAAGCCAGAGUCGGCCGAGAGAGUGGGUGAGAGUAGUGGAAAGUGGAAAAUUUGCU (.(((((((((((.(((((....))))).((((((...))))))...)))))))...))))).....((..(((.....((.(....).))....)))..)) ( -25.20) >DroYak_CAF1 12149 102 + 1 GUGCCGCUGGCUUGGCAUAUGAUUGUGUAAUUGAUUUGAUCAAUAGAAAGCCGGAGUCGGCGGCGAGUGAGGGUGAAAGUAGUGGAAAGUGGAAAAUUUGCU .(((((((((((((((((((....)))).((((((...)))))).....))).))))))))))))...............((..((...(....).))..)) ( -29.50) >consensus GUGCCGCUGGCUUGGCAUAUGAUUGUGUAAUUGAUUUGAUCAAUAGAAAGCCAGAGUCGGCCGAGAGAGUGGGUGAGAGUAGUGGAAAGUGGAAAAUUUGCU ..(((((((((((.(((((....))))).((((((...))))))...)))))))...))))..........................((..((...))..)) (-21.16 = -21.47 + 0.31)



| Location | 17,982,086 – 17,982,178 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -17.06 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.31 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17982086 92 - 22224390 AGCAAAUUUUCCACUUU----------CACCCGCACUCUCGGCCGACUCUGGCUUUCUAUUGAUCAAAUCAAUUACACAAUCAUAUGCCAAGCCAGCGGCAC .................----------..............((((...(((((((..(((((((...............)))).)))..))))))))))).. ( -17.06) >DroSec_CAF1 12994 100 - 1 AGCAAAUUUUCCACUUUCCACUACUCUCACCCACUCUCUCGGCCGACUCUGGCU-UCUA-UGAUCAAAUCAAUUACACAAUCAUAUGCCAAGCCAGCGGCAC .........................................((((...((((((-(.((-((((...............))))))....))))))))))).. ( -18.06) >DroSim_CAF1 12713 102 - 1 AGCAAAUUUUCCACUUUCCACUACUCUCACCCACUCUCUCGGCCGACUCUGGCUUUCUAUUGAUCAAAUCAAUUACACAAUCAUAUGCCAAGCCAGCGGCAC .........................................((((...(((((((..(((((((...............)))).)))..))))))))))).. ( -17.06) >DroYak_CAF1 12149 102 - 1 AGCAAAUUUUCCACUUUCCACUACUUUCACCCUCACUCGCCGCCGACUCCGGCUUUCUAUUGAUCAAAUCAAUUACACAAUCAUAUGCCAAGCCAGCGGCAC ......................................(((((.(....)(((((..(((((((...............)))).)))..))))).))))).. ( -16.06) >consensus AGCAAAUUUUCCACUUUCCACUACUCUCACCCACACUCUCGGCCGACUCUGGCUUUCUAUUGAUCAAAUCAAUUACACAAUCAUAUGCCAAGCCAGCGGCAC .........................................((((...(((((((.....((((...............))))......))))))))))).. (-14.81 = -15.31 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:56 2006