
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,975,479 – 1,975,578 |
| Length | 99 |
| Max. P | 0.999462 |

| Location | 1,975,479 – 1,975,578 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.45 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1975479 99 + 22224390 UGAAUGGCAGCAAUUUGCUGUUGCAACAUUUGUUGUUGAAGGAUGUGCUGUUUCUGAAACAGUUGUAGCUGC---------------------UGUUGCUGCUUGUCGACUCCUGCCGCA .((..(((((((((..((.((((((((......((((..((((........))))..)))))))))))).))---------------------.)))))))))..))............. ( -31.90) >DroSec_CAF1 8354 87 + 1 UGAAUGGCAGCAA------------ACAUUUGUUGCUGCAGGAUUUGCUGUUUUUGAAUCAGUUGUAGCUGC---------------------UGUUGCUGCUUGUCGACUCCUGCCGCA .....((((((((------------....))))))))((((((...((((.........)))).(((((...---------------------....)))))........)))))).... ( -26.40) >DroYak_CAF1 8561 120 + 1 UGAAUGGCAGCAAUCUGCUGUUGCAACAUUUGCUGCUGUAGGAUUUGCUGUUUCUGGAUCAGUUGCAACUGCUGUUUCUCAAUCAGUUGUAACUGUUGCUGCUUGUCCACUCCUGCCGUA ...((((((((((..(((....)))....)))))))))).((((..((.((........((((((((((((.((.....))..))))))))))))..)).))..))))............ ( -40.50) >consensus UGAAUGGCAGCAAU_UGCUGUUGCAACAUUUGUUGCUGAAGGAUUUGCUGUUUCUGAAUCAGUUGUAGCUGC_____________________UGUUGCUGCUUGUCGACUCCUGCCGCA .....((((((((..(((....)))....))))))))((((((...((((.........)))).(((((.((......................)).)))))........)))))).... (-22.24 = -22.38 + 0.15)

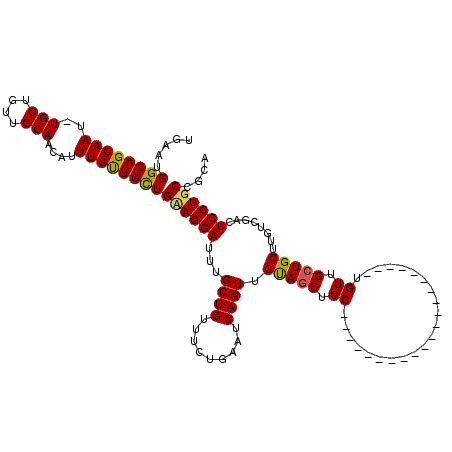
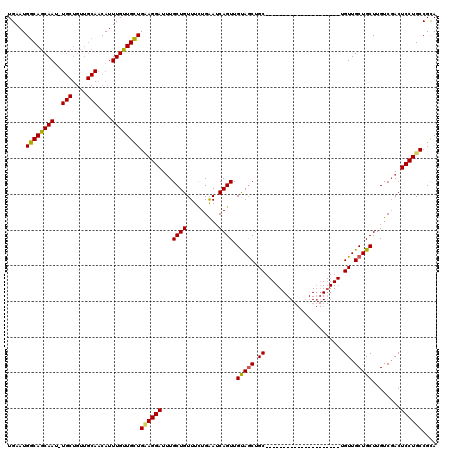
| Location | 1,975,479 – 1,975,578 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.45 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1975479 99 - 22224390 UGCGGCAGGAGUCGACAAGCAGCAACA---------------------GCAGCUACAACUGUUUCAGAAACAGCACAUCCUUCAACAACAAAUGUUGCAACAGCAAAUUGCUGCCAUUCA ((((((....)))).)).(((((((..---------------------((........(((((.....))))).........(((((.....))))).....))...)))))))...... ( -22.40) >DroSec_CAF1 8354 87 - 1 UGCGGCAGGAGUCGACAAGCAGCAACA---------------------GCAGCUACAACUGAUUCAAAAACAGCAAAUCCUGCAGCAACAAAUGU------------UUGCUGCCAUUCA ...(((((((((((...(((.((....---------------------)).))).....)))))).......(((.....)))............------------...)))))..... ( -21.00) >DroYak_CAF1 8561 120 - 1 UACGGCAGGAGUGGACAAGCAGCAACAGUUACAACUGAUUGAGAAACAGCAGUUGCAACUGAUCCAGAAACAGCAAAUCCUACAGCAGCAAAUGUUGCAACAGCAGAUUGCUGCCAUUCA ........((((((....((((((.(((((.((((((.(((.....))))))))).)))))...........((..........))......))))))..((((.....)))))))))). ( -35.90) >consensus UGCGGCAGGAGUCGACAAGCAGCAACA_____________________GCAGCUACAACUGAUUCAGAAACAGCAAAUCCUACAGCAACAAAUGUUGCAACAGCA_AUUGCUGCCAUUCA ..((((....))))....(((((((........................(((......))).....................(((((.....)))))..........)))))))...... (-15.19 = -15.97 + 0.78)

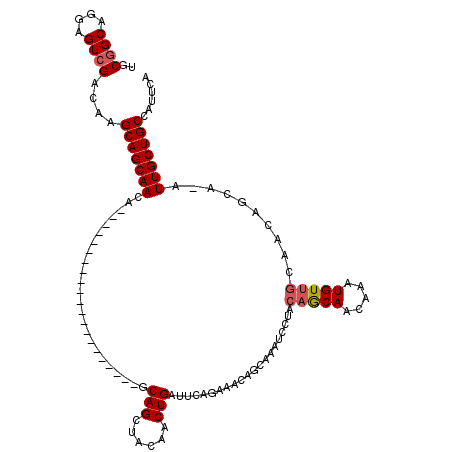
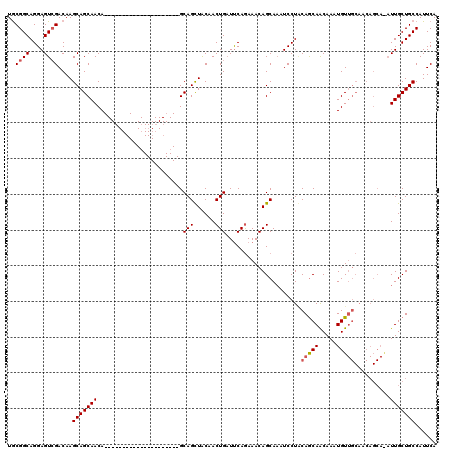
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:56 2006