| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,977,133 – 17,977,244 |
| Length | 111 |
| Max. P | 0.733358 |

| Location | 17,977,133 – 17,977,244 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -19.45 |
| Energy contribution | -20.21 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17977133 111 + 22224390 UUUGCAUUUGCGCAGGCAGCCGCAAAACACCUCGUCCCCAAAAGCUCCCAACCCAA---------ACAGCCAUUUCUCUUUGUUGUCGACGCCGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(..((((..(((..(((..........---------..)))((........)))))..))))..)..))))))....)))))..))))).. ( -23.40) >DroSec_CAF1 8017 107 + 1 UUUGCAUUUGCGCAGGCAGCCGCAAAACACCUCGUCCCCAGCCGCCCC----CCAA---------ACAGCCAUUUCUCUUUGUUGUCGACGCCGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(..((((..((((......----....---------................))))..))))..)..))))))....)))))..))))).. ( -24.57) >DroSim_CAF1 7723 107 + 1 UUUGCAUUUGCGCAGGCAGCCGCAAAACACCUCGUCCCCAACCGCCCC----CCAA---------ACAGCCAUUUCUCUUUGUUGUCGACGCCGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(..((((..((((......----....---------................))))..))))..)..))))))....)))))..))))).. ( -24.57) >DroEre_CAF1 9369 116 + 1 UUUGCAUUUGCGCAGGCAGCCGCAAAACACUUCGCCCCCAAACAGCCC----CCAACCGCCCCCCACAACCAUUUCUCUUUGUUGUCGACGACGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(.(((..........((..----......))...(.(((((...........))))).).))).)..))))))....)))))..))))).. ( -22.40) >DroYak_CAF1 7013 107 + 1 UUUGCAUUUGCGCAGGCAGCCGCAAAACACCUCGCCCCCCAUUCGCCU----CCAA---------AGAACCAUUUCUCUUUGUUGUCGACGCCGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(..((........(((.(.----.(((---------(((........))))))..).)))))..)..))))))....)))))..))))).. ( -24.20) >consensus UUUGCAUUUGCGCAGGCAGCCGCAAAACACCUCGUCCCCAAACGCCCC____CCAA_________ACAGCCAUUUCUCUUUGUUGUCGACGCCGCUUUUUGCUUUAUGCUUACGCGCAUU ........((((((((((...((((((..(..((((..((((.......................................))))..))))..)..))))))....)))))..))))).. (-19.45 = -20.21 + 0.76)


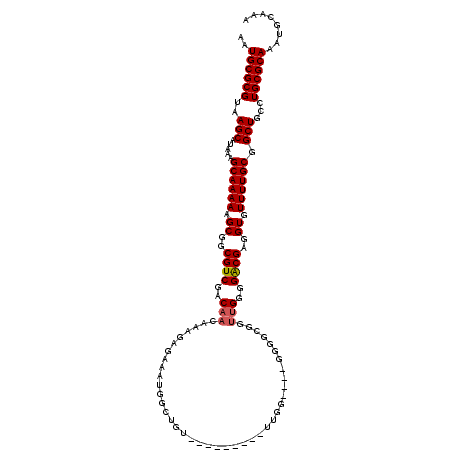
| Location | 17,977,133 – 17,977,244 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17977133 111 - 22224390 AAUGCGCGUAAGCAUAAAGCAAAAAGCGGCGUCGACAACAAAGAGAAAUGGCUGU---------UUGGGUUGGGAGCUUUUGGGGACGAGGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..((((((..(((.....((((((.(((((.(((((..((((.((......)).)---------))).)))))..)))((((....)))))).)))))).)))...))))))........ ( -34.90) >DroSec_CAF1 8017 107 - 1 AAUGCGCGUAAGCAUAAAGCAAAAAGCGGCGUCGACAACAAAGAGAAAUGGCUGU---------UUGG----GGGGCGGCUGGGGACGAGGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..(((((....)).....((...(((((((.((...........))....)))))---------)).(----.((((((((((((((.....))))).))))))))).)))....))).. ( -36.00) >DroSim_CAF1 7723 107 - 1 AAUGCGCGUAAGCAUAAAGCAAAAAGCGGCGUCGACAACAAAGAGAAAUGGCUGU---------UUGG----GGGGCGGUUGGGGACGAGGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..((((((..(((.....((((((.((..((((..((((....(....).(((.(---------....----).))).))))..))))..)).)))))).)))...))))))........ ( -34.50) >DroEre_CAF1 9369 116 - 1 AAUGCGCGUAAGCAUAAAGCAAAAAGCGUCGUCGACAACAAAGAGAAAUGGUUGUGGGGGGCGGUUGG----GGGCUGUUUGGGGGCGAAGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..(((((....)).....((......((((.((.(((((...........))))).)).))))((.((----.((((((..(((.((...)).))).)))))).)).))))....))).. ( -37.90) >DroYak_CAF1 7013 107 - 1 AAUGCGCGUAAGCAUAAAGCAAAAAGCGGCGUCGACAACAAAGAGAAAUGGUUCU---------UUGG----AGGCGAAUGGGGGGCGAGGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..((((((..(((.....((((((.((..((((..(..(((((((......))))---------))).----.........)..))))..)).)))))).)))...))))))........ ( -31.30) >consensus AAUGCGCGUAAGCAUAAAGCAAAAAGCGGCGUCGACAACAAAGAGAAAUGGCUGU_________UUGG____GGGGCGGUUGGGGACGAGGUGUUUUGCGGCUGCCUGCGCAAAUGCAAA ..((((((..(((.....((((((.((..((((..(((.........................................)))..))))..)).)))))).)))...))))))........ (-25.26 = -25.42 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:50 2006