
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,975,653 – 17,975,883 |
| Length | 230 |
| Max. P | 0.547795 |

| Location | 17,975,653 – 17,975,765 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17975653 112 + 22224390 UGGGGUUCUUCGGCAGUCGUGGAGGCGCCACCAACACGACAAAGUUGUCGCUGGCGAAUCUGGAAUUAUUAUUGAAUGAACUUGCACCAUAUCCGCC--------GUCCAUUUCGCCAUA ((((((.((((.((....)).)))).))).)))...(((((....))))).(((((((..((((.(((((....)))))....((.........)).--------.)))).))))))).. ( -33.50) >DroSec_CAF1 6593 103 + 1 ---------GCGGCAGUCGUGGAGGCGCCACCAACACGACAAAGUUGUCGCUGGCGAAUCUGGAAUUAUUAGUGAAUGAACUUGCGCCAUAUCCGCC--------GUCCAUUUCGCCAUA ---------((((((((((((..((.....))..)))))).....))))))(((((((..((((......(((......))).(((.......))).--------.)))).))))))).. ( -35.70) >DroSim_CAF1 6284 103 + 1 ---------GCGGCAGUCGUGGAGGCGCCACCAACACGACAAAGUUGUCGCUGGCGAAUCUGGAAUUAUUAUUGAAUGAACUUGCGCCAUAUCCGCC--------GUCCAUUUCGCCAUA ---------((((((((((((..((.....))..)))))).....))))))(((((((..((((.(((((....)))))....(((.......))).--------.)))).))))))).. ( -35.50) >DroEre_CAF1 8214 100 + 1 -----------GGCAGUCGCGAAGGCGCCAGCAACACGACAAAGUUGUCGCAGGCGAAUC-GAAAUUAUUAUUGAAUGAACUAGCGCCAUAUCCGCA--------GUCCAUUUCGCCAUA -----------.((.(.(((....))).).))....(((((....)))))..((((((((-((........))))(((.(((.(((.......))))--------)).)))))))))... ( -28.20) >DroYak_CAF1 5610 108 + 1 -----------GGCAGCCGUGGAGGCGCCACCAACACGACAAAGUUGUCGCUGGCGAAUU-GGAAUUAUUAUUGAAUGAACAUGCGCCACAUCCGCAGAUCCACCGUCCACUUCGCCAUA -----------(((.(((.....)))))).......(((((....))))).(((((((.(-(((.(((((....)))))...((((.......)))).........)))).))))))).. ( -34.90) >consensus __________CGGCAGUCGUGGAGGCGCCACCAACACGACAAAGUUGUCGCUGGCGAAUCUGGAAUUAUUAUUGAAUGAACUUGCGCCAUAUCCGCC________GUCCAUUUCGCCAUA ...........(((.(((.....)))))).......(((((....)))))..((((((...(((.(((((....)))))....(((.......)))..........)))..))))))... (-26.02 = -26.26 + 0.24)

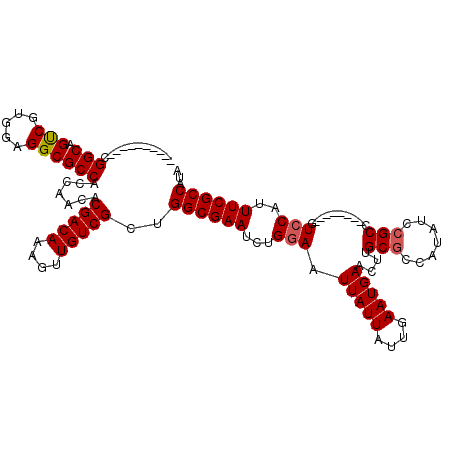

| Location | 17,975,765 – 17,975,883 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.94 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17975765 118 - 22224390 AUACGCGCCUGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGGCAGCGGCGAGUCCGAAGUUUUUGGAUAUCCCCA--CCUCAUCUUGGCUGCUUUAUUGGCGGCGCUUCAAUAAUUGAU .........(((((.(((((((((((.........(....)((((((((.((((((((.....)))))).)).)).--.(.......)))))))))))))))))).)))))......... ( -36.20) >DroSec_CAF1 6696 116 - 1 AUACGCGCCUGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGGCAGCGGCGAGGCCAGGGUUUUUG-GUAUC-CCA--CCUCGUCCUGGCUGCUUUAUUGGCGGCGCUUCAAUAAUUGAU .........(((((.(((((((((((.........(....)(((((((((((((...(((......-....)-)).--)))))))...))))))))))))))))).)))))......... ( -42.80) >DroSim_CAF1 6387 117 - 1 AUACGCGCCUGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGGCAGCGGCGAGGCCAGGGUUUUUGGGUAUC-CCA--CCUCGUCCUGGCUGCUUUAUUGGCGGCGCUUCAAUAAUUGAU .........(((((.(((((((((((......(..(....)..)((((((.(((.(..(((....(((...)-)))--))..).))).))))))))))))))))).)))))......... ( -43.60) >DroEre_CAF1 8314 116 - 1 AUACGCGCCUGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGGCAGCGGCGAGGCCGUAGUUUUUGGGUAUC-CCA--ACUCAUC-UGGCUGCUUUAUGGGCGGCGCUUCAAUAAUUGAU ....(((((.((....))..............(..((...((((((((((...))).(((...(((((...)-)))--).....)-)))))))))..))..))))))............. ( -34.10) >DroYak_CAF1 5718 118 - 1 AUACGCGCCAGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGCCAGCGGCGAGGCCGAAGUUUUUGGUUAUA-CCAAAACUCAUC-UGGCUGCUUUAUUGGCGGCGCUUCAAUAAUUGAU ....(((((.((....))((((((((......(..(....)..)((((((.(((..((.(((((.((.....-))))))))).))-).)))))))))))))))))))............. ( -38.80) >consensus AUACGCGCCUGGGGAGCCGUCAAUAAUUAAUCGAACAAAUGGGCAGCGGCGAGGCCGAAGUUUUUGGGUAUC_CCA__CCUCAUC_UGGCUGCUUUAUUGGCGGCGCUUCAAUAAUUGAU ....(((((.((....))((((((((......(..(....)..)((((((.(((....((....(((......)))...))...))).)))))))))))))))))))............. (-31.26 = -31.94 + 0.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:48 2006