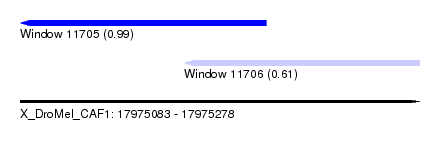
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,975,083 – 17,975,278 |
| Length | 195 |
| Max. P | 0.992608 |

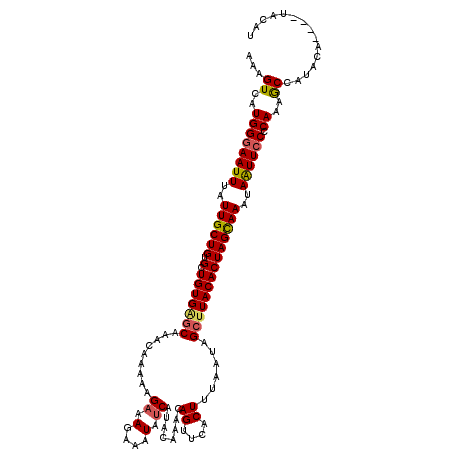
| Location | 17,975,083 – 17,975,203 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17975083 120 - 22224390 AAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAAAAGAAAGAAAUAUCAUACAAACAGUUCACUUUAAUAGCUUACACUAGCAAAUAAUUCCCCAAAGCCAUAAAUAUGUACAU ........(((((((..((((((.(.(((((((.........((.(....).))........((....))......))))))))))))))..)))))))..................... ( -23.80) >DroSec_CAF1 5984 112 - 1 AAAGUCAUGGGAAUUUAUUGCUGUGCUGUGGGCAAACAAAAAGAAAGAAAUAUCAUACAAACAGUUCACUUUAAUAGCUUACACUAGCAAAUAAUUCCCCAAAGCCAUACA--------U ........(((((((..((((((.(.(((((((.........((.(....).))........((....))......))))))))))))))..)))))))............--------. ( -23.00) >DroSim_CAF1 5673 116 - 1 AAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAAAAGAAAGAAAUAUCAUACAAACAGUUCACUUUAAUAGCUUACACUAGCAAAUAAUUCCCCAAAGCCAUACAU----ACAU ........(((((((..((((((.(.(((((((.........((.(....).))........((....))......))))))))))))))..))))))).............----.... ( -23.80) >DroEre_CAF1 7683 117 - 1 AAAGUCAUGGAAAUUUAUUGCUGUGCUGUGAGCAAACAAAAUGAAACAAAUACCAUACAAACAGUUCACUUUAAUGGCUUACACUAGUAAAUAGUUCC-CAAAACCAUAUA--AGUUCAC ...((..(((.((((..((((((.(.(((((((....(((.((((...................)))).)))....))))))))))))))..)))).)-))..))......--....... ( -18.21) >DroYak_CAF1 5115 117 - 1 AAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAACAAACAAAAAGAAAGAAAUAUCAUACAAACAGUUCACUUUAAUAGCGUACACUAGCAAAUAAUUCC-CAAAGCCAUAUA--CAUACAU ...((..((((((((..((((((((((((((((.........((.(....).)).........))))))......)))).....))))))..))))))-))..))......--....... ( -23.67) >consensus AAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAAAAGAAAGAAAUAUCAUACAAACAGUUCACUUUAAUAGCUUACACUAGCAAAUAAUUCCCCAAAGCCAUACA____UACAU ...((..((((((((..((((((.(.(((((((.........((.(....).))........((....))......))))))))))))))..)))))).))..))............... (-18.02 = -18.22 + 0.20)


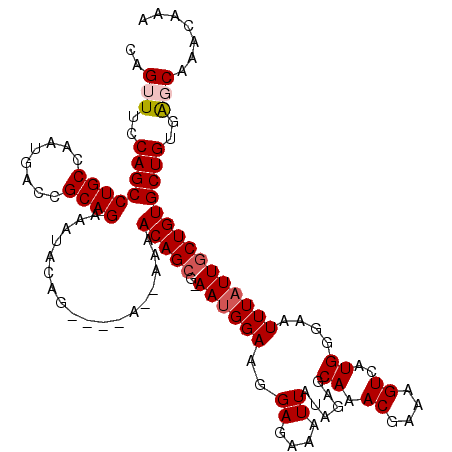
| Location | 17,975,163 – 17,975,278 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17975163 115 - 22224390 CAGUUUCCAGCCUGCCAAUGACCGCAGAAAUACAA----AAAAAAACAGCG-AAUGGAAGGAGAAAUUAUAGAGCAAACGAAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAA ..(((..((((((((........))))........----......(((((.-((((((..((....))......((.((....))..))....)))))))))))))))..)))....... ( -24.60) >DroSec_CAF1 6056 113 - 1 CAGUUUCCAGCCUGCCAAUGACCGCAGAAAUACAG----A--AAAACAGCG-AAUGGAAGGAGAAAUUAUAGAGCAAACGAAAGUCAUGGGAAUUUAUUGCUGUGCUGUGGGCAAACAAA ..((((((...((((........))))...(((((----.--...(((((.-((((((..((....))......((.((....))..))....))))))))))).))))))).))))... ( -25.50) >DroSim_CAF1 5749 113 - 1 CAGUUUCCAGCCUGCCAAUGACCGCAGAAAUACAG----A--AAAACAGCG-AAUGGAAGGAGAAAUUAUAGAGCAAACGAAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAA ..(((..((((((((........))))........----.--...(((((.-((((((..((....))......((.((....))..))....)))))))))))))))..)))....... ( -24.60) >DroEre_CAF1 7760 117 - 1 CGGAUUCCAGCCUGCGAAUGACCGCAGAAAUAAAGCGAAA--AAAACAGCG-AAUGGAAGGAGAAAUUAUAGAGCAAACGAAAGUCAUGGAAAUUUAUUGCUGUGCUGUGAGCAAACAAA ..(.((((((((((((......))))).............--...(((((.-((((((..((....))......((.((....))..))....))))))))))))))).))))....... ( -24.80) >DroYak_CAF1 5192 114 - 1 CAGAUUCCAGCCUGCCAAUGACCGCAGAAAUACAG----A--AAAACAGAAAAAAGGAAGGAGAAAUUAUAGCGCAAACGAAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAACAAACAAA ....((((...((((........)))).......(----.--....)........)))).......(((((((((..((....))...((.((....)).)))))))))))......... ( -18.80) >consensus CAGUUUCCAGCCUGCCAAUGACCGCAGAAAUACAG____A__AAAACAGCG_AAUGGAAGGAGAAAUUAUAGAGCAAACGAAAGUCAUGGGAAUUUAUUGCUGUGCUGUGAGCAAACAAA ..(((..((((((((........))))..................(((((..((((((..((....))......((.((....))..))....)))))))))))))))..)))....... (-20.08 = -20.72 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:46 2006