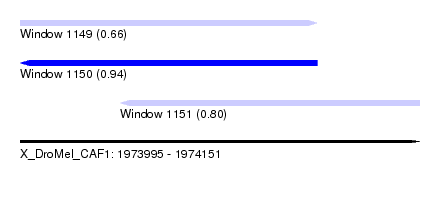
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,973,995 – 1,974,151 |
| Length | 156 |
| Max. P | 0.937904 |

| Location | 1,973,995 – 1,974,111 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -35.90 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973995 116 + 22224390 ACGU-GGUGGUUGUGGUUGUGCUGGUGCAACUUGUGGUAAAGGUGCCAGAACUGGUAGUGGUUGUGCUAGCGGUUGAGACAGCCGU---GCUGGCUGUCGUUCUGGCUACUCCAAUCCCA ...(-((.(((((..((.(((((((..(((((.........(.(((((....))))).))))))..)))))......(((((((..---...)))))))......)).))..)))))))) ( -46.20) >DroSec_CAF1 6878 120 + 1 ACGUAGGUGGUUGUUGUUGUGCUGGUGCAACUUGUGGUAAAGGUGCCAGAACUGGUAGUGGUUGUGCUAGCGGUUGAGACAGCCGUGGUGCUGGCUGUCGUUCUGGCUACUCCAAUCCCG .....((.(((((..((.(((((((..(((((.........(.(((((....))))).))))))..)))))......((((((((......))))))))......)).))..))))))). ( -45.40) >DroYak_CAF1 7110 113 + 1 ACGU-GGUGGUUGUGGUUGUGCUGGUACAACUUGUAGUAAAGGUGCCAGAACUGAU---GGUUGUG---GUGGUUGAGCCAGUAGUGGUCGUAGCUGUCGUUUUGGCUACUCCAAUUCCA ....-..(((...(((...(((((((.(((((............((((.(((....---.))).))---))))))).)))))))(((((((.(((....))).))))))).)))...))) ( -36.70) >consensus ACGU_GGUGGUUGUGGUUGUGCUGGUGCAACUUGUGGUAAAGGUGCCAGAACUGGUAGUGGUUGUGCUAGCGGUUGAGACAGCCGUGGUGCUGGCUGUCGUUCUGGCUACUCCAAUCCCA .....((.(((((..((((((....))))))..........(((((((((((.(.((((......)))).)......((((((((......)))))))))))))))).))).))))))). (-35.90 = -36.30 + 0.40)

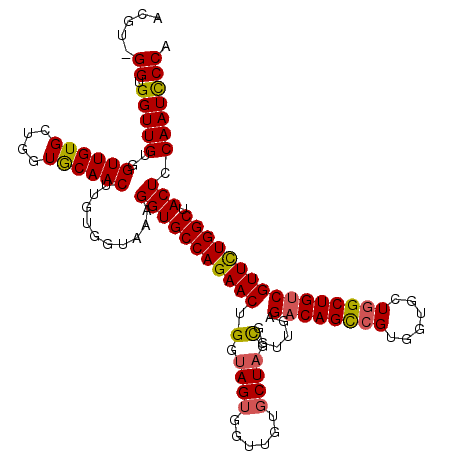
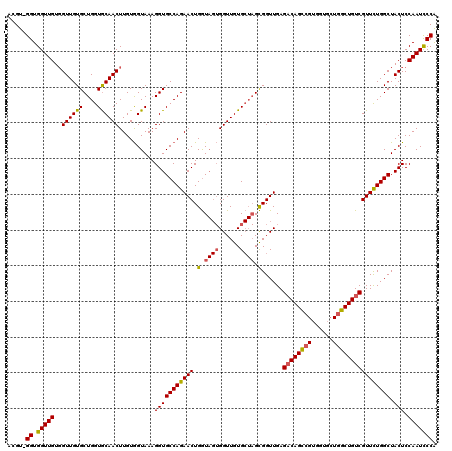
| Location | 1,973,995 – 1,974,111 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973995 116 - 22224390 UGGGAUUGGAGUAGCCAGAACGACAGCCAGC---ACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCUUUACCACAAGUUGCACCAGCACAACCACAACCACC-ACGU (((...(((....(((((((((((((((...---..)))))))......(.(((........))).).)))))))).............((.((....))))..)))...)))..-.... ( -36.20) >DroSec_CAF1 6878 120 - 1 CGGGAUUGGAGUAGCCAGAACGACAGCCAGCACCACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCUUUACCACAAGUUGCACCAGCACAACAACAACCACCUACGU .(((..(((((..(((((((((((((((........)))))))......(.(((........))).).))))))))..)))))......((((........))))........))).... ( -36.30) >DroYak_CAF1 7110 113 - 1 UGGAAUUGGAGUAGCCAAAACGACAGCUACGACCACUACUGGCUCAACCAC---CACAACC---AUCAGUUCUGGCACCUUUACUACAAGUUGUACCAGCACAACCACAACCACC-ACGU (((...(((.(((((..........)))))........((((..((((..(---((.(((.---....))).)))..............))))..)))).....)))...)))..-.... ( -20.09) >consensus UGGGAUUGGAGUAGCCAGAACGACAGCCAGCACCACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCUUUACCACAAGUUGCACCAGCACAACCACAACCACC_ACGU .((...(((((..(((((((((((((((........)))))))......(....).............))))))))..)))))......((((........)))).....))........ (-25.23 = -26.90 + 1.67)

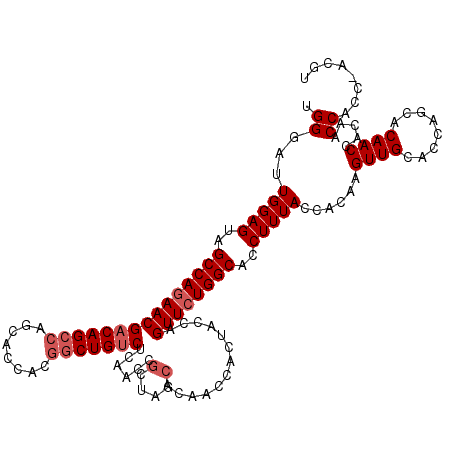
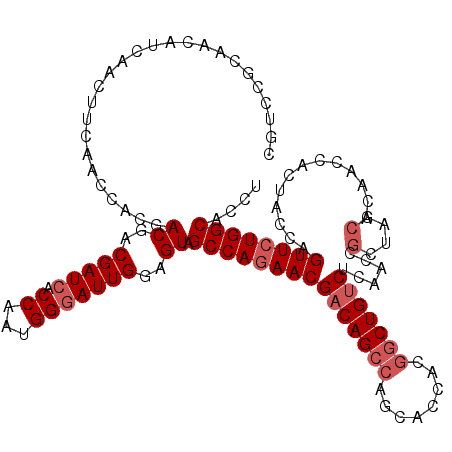
| Location | 1,974,034 – 1,974,151 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -23.33 |
| Energy contribution | -25.33 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1974034 117 - 22224390 CGUCCGCAACAUCAACAUCAACCACGACAACGAUCACCAAUGGGAUUGGAGUAGCCAGAACGACAGCCAGC---ACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCU ..........................((..(((((.((...)))))))..)).(((((((((((((((...---..)))))))......(.(((........))).).)))))))).... ( -34.70) >DroSec_CAF1 6918 120 - 1 CGUCCGCAACAUCAACUUCAACCACGACGACGAUCACCAACGGGAUUGGAGUAGCCAGAACGACAGCCAGCACCACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCU ..............(((((((((.((..(........)..)))).))))))).(((((((((((((((........)))))))......(.(((........))).).)))))))).... ( -37.80) >DroYak_CAF1 7149 114 - 1 CGUCCGCAACAUCAACUUCUACCACGACGACGAUCACCAAUGGAAUUGGAGUAGCCAAAACGACAGCUACGACCACUACUGGCUCAACCAC---CACAACC---AUCAGUUCUGGCACCU .(((((.((((((..(.((......)).)..))).....((((...(((.(((((..........)))))(((((....))).)).....)---))...))---))..))).))).)).. ( -19.70) >consensus CGUCCGCAACAUCAACUUCAACCACGACGACGAUCACCAAUGGGAUUGGAGUAGCCAGAACGACAGCCAGCACCACGGCUGUCUCAACCGCUAGCACAACCACUACCAGUUCUGGCACCU ..........................((..(((((.((...)))))))..)).(((((((((((((((........)))))))......(....).............)))))))).... (-23.33 = -25.33 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:54 2006