
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,971,096 – 17,971,417 |
| Length | 321 |
| Max. P | 0.964420 |

| Location | 17,971,096 – 17,971,194 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971096 98 + 22224390 GGUCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGU-GUGUGU------------AAAAUCAUGUAAA ....((((((((((((.......)).))))))...(((((......))))).---------...((((.((((.........))))))-))))))------------............. ( -25.10) >DroSec_CAF1 1819 110 + 1 GGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGU-GUGUGUCCGCAGCACCGUUUAAUCAUGUAAA ((((.....(((......)))......))))....(((((......))))).---------.......(((((((((((.((((.(((-(......))))))))))))))).)))).... ( -32.50) >DroSim_CAF1 1182 110 + 1 GGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGU-GUGUGUCCGCAGCACCGUUUAAUCAUGUAAA ((((.....(((......)))......))))....(((((......))))).---------.......(((((((((((.((((.(((-(......))))))))))))))).)))).... ( -32.50) >DroEre_CAF1 1927 116 + 1 GGCCACACUGGCCUGCACGCUUAACAGGGGCUAACUGCUGAUUUAACAGCGAAAAGCCAAAAUAACACUAUGUUAAAUGUGUGCAUGU-GUGUGCCCG---CACUGUUUUAUCAUGUAAA ((((.....))))(((((((((((((..((((...(((((......)))))...))))............))))))..))))))).((-(((....))---)))................ ( -35.94) >DroYak_CAF1 1322 117 + 1 GGCCACACUGAUAUGCACGCUUAACAGGGGCAAACUGCUGAUUUAACAGCGAAAAGCCAGAAUAACACUAUGUUAAAUGUGUCUAUGUGGUGUGCCCA---CACCGUUUAAUCAUGUAAA ((.((((((((((.(((((.((((((..(((....(((((......)))))....)))............)))))).))))).))).))))))).)).---................... ( -31.14) >consensus GGCCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGA_________ACAACACUAUGUUAAAUGUGUGCAUGU_GUGUGUCCG___CACCGUUUAAUCAUGUAAA ((.(((((......((((((((((((((......))((((......))))....................))))))..)))))).....))))).))....................... (-22.04 = -22.12 + 0.08)



| Location | 17,971,136 – 17,971,229 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971136 93 + 22224390 AUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGUGUGUGU------------AAAAUCAUGUAAAAUCGACCG-AACGUCGAUGGGACAAACAAAAA---AAAA ............---------(((.((((((((.........))))).))))))------------.......(((...((((((..-...))))))...)))........---.... ( -16.40) >DroSec_CAF1 1859 108 + 1 AUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGUGUGUGUCCGCAGCACCGUUUAAUCAUGUAAAAUCGACAG-AACGUCGAUAGGACAAACAAAAAAAAAAAA ............---------..........((((((((.((((.((((......))))))))))))))))..(((...((((((..-...))))))...)))............... ( -24.10) >DroSim_CAF1 1222 108 + 1 AUUUAACAGCGA---------ACAACACUAUGUUAAAUGUGUGCAUGUGUGUGUCCGCAGCACCGUUUAAUCAUGUAAAAUCGACAG-AACGUCGAUAGGACAAACAAAAAAAAAAAA ............---------..........((((((((.((((.((((......))))))))))))))))..(((...((((((..-...))))))...)))............... ( -24.10) >DroEre_CAF1 1967 111 + 1 AUUUAACAGCGAAAAGCCAAAAUAACACUAUGUUAAAUGUGUGCAUGUGUGUGCCCG---CACUGUUUUAUCAUGUAAAAUCGUCAAUAAAGUCGAUAGGACAA----AAAAAAAAAA ....(((((.............((((.....))))..((((.((((....)))).))---)))))))......(((...((((.(......).))))...))).----.......... ( -17.70) >consensus AUUUAACAGCGA_________ACAACACUAUGUUAAAUGUGUGCAUGUGUGUGUCCG___CACCGUUUAAUCAUGUAAAAUCGACAG_AACGUCGAUAGGACAAACAAAAAAAAAAAA ........................(((((((((.........))))).)))).....................(((...((((((......))))))...)))............... (-14.32 = -14.58 + 0.25)


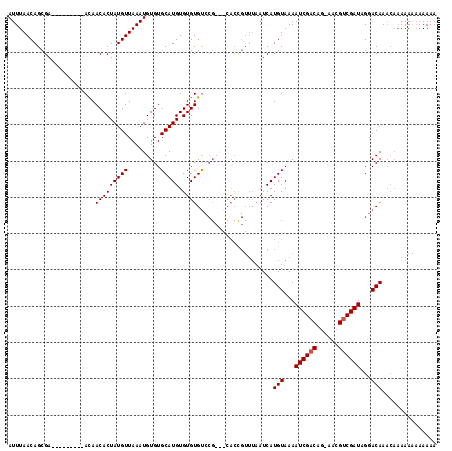
| Location | 17,971,167 – 17,971,268 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971167 101 + 22224390 GUGCAUGUGUGUGU------------AAAAUCAUGUAAAAUCGACCG-AACGUCGAUGGGACAAACAAAAA---AAAACAACGAAAUUAUGUACACAAAUGCAUUUUUCU-UUAGGGC (((((((((((..(------------((..((.......((((((..-...)))))).(......).....---........))..)))..))))))..)))))......-....... ( -20.70) >DroSec_CAF1 1890 116 + 1 GUGCAUGUGUGUGUCCGCAGCACCGUUUAAUCAUGUAAAAUCGACAG-AACGUCGAUAGGACAAACAAAAAAAAAAAAAAACGCAAUUAUGUACACAAAUGCAUUUUUCU-UUAGGGC ((((((.((((((((((((..............)))...((((((..-...)))))).))))....................(((....)))))))).))))))......-....... ( -25.34) >DroSim_CAF1 1253 116 + 1 GUGCAUGUGUGUGUCCGCAGCACCGUUUAAUCAUGUAAAAUCGACAG-AACGUCGAUAGGACAAACAAAAAAAAAAAACAACGCAAUUAUGUACACAAAUGCAUUUUUCU-UUAGGGC ((((((.((((((((((((..............)))...((((((..-...)))))).))))....................(((....)))))))).))))))......-....... ( -25.34) >DroEre_CAF1 2007 108 + 1 GUGCAUGUGUGUGCCCG---CACUGUUUUAUCAUGUAAAAUCGUCAAUAAAGUCGAUAGGACAA----AAAAAAAAAAC---AAAAUUAUGUACACAAAUGCACUUAUCUUUUAGAGC ((((((.(((((((...---...((((((....(((...((((.(......).))))...))).----......)))))---).......))))))).)))))).............. ( -24.72) >consensus GUGCAUGUGUGUGUCCG___CACCGUUUAAUCAUGUAAAAUCGACAG_AACGUCGAUAGGACAAACAAAAAAAAAAAACAACGAAAUUAUGUACACAAAUGCAUUUUUCU_UUAGGGC ((((((.((((((..((......))........(((...((((((......))))))...)))............................)))))).)))))).............. (-19.04 = -19.10 + 0.06)



| Location | 17,971,229 – 17,971,348 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971229 119 + 22224390 CAACGAAAUUAUGUACACAAAUGCAUUUUUCUUUAGGGCAGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACA ....((((..(((((......))))).)))).....((((((((((((((....)))))))))((......)).((((((((((((......))))))......)))))).)))))... ( -31.90) >DroSec_CAF1 1967 119 + 1 AAACGCAAUUAUGUACACAAAUGCAUUUUUCUUUAGGGCAGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACA ....(((((....(((((............(((.((((((((....)))))))).)))((((((((........))....((((((......))))))))))))))))).))))).... ( -31.50) >DroSim_CAF1 1330 119 + 1 CAACGCAAUUAUGUACACAAAUGCAUUUUUCUUUAGGGCAGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUAUGUAUUGCCACA ....(((((.((((((..............(((.((((((((....)))))))).)))((((((((........))....((((((......))))))))))))))))))))))).... ( -31.50) >consensus CAACGCAAUUAUGUACACAAAUGCAUUUUUCUUUAGGGCAGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACA ....(((((.(..(((..............(((.((((((((....)))))))).)))((((((((........))....((((((......)))))))))))))))..)))))).... (-28.95 = -29.07 + 0.11)



| Location | 17,971,229 – 17,971,348 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -33.25 |
| Energy contribution | -33.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971229 119 - 22224390 UGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCUGCCCUAAAGAAAAAUGCAUUUGUGUACAUAAUUUCGUUG .((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))......((((.........(((.......)))....((((....)))).....)))).... ( -31.30) >DroSec_CAF1 1967 119 - 1 UGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCUGCCCUAAAGAAAAAUGCAUUUGUGUACAUAAUUGCGUUU .((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))...................(((.......))).(((((((.((((....)))).))))))) ( -36.10) >DroSim_CAF1 1330 119 - 1 UGUGGCAAUACAUACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCUGCCCUAAAGAAAAAUGCAUUUGUGUACAUAAUUGCGUUG .((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))...................(((.......)))..((((((.((((....)))).)))))). ( -34.40) >consensus UGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCUGCCCUAAAGAAAAAUGCAUUUGUGUACAUAAUUGCGUUG .((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))...................(((.......)))..((((((.((((....)))).)))))). (-33.25 = -33.37 + 0.11)



| Location | 17,971,268 – 17,971,378 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -26.55 |
| Energy contribution | -26.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971268 110 + 22224390 AGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACACGUACACAAAGGCACAGUCGAAAAGC ..(((((((((....)))))))))(((.....((.......((((((......))))))...(((((((((........)))))))))......))....)))....... ( -28.50) >DroSec_CAF1 2006 110 + 1 AGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAAAGC .((((.((((((.......((((((((........))....((((((......))))))))))))((((((......))))))......))))))....))))....... ( -28.10) >DroSim_CAF1 1369 110 + 1 AGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUAUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAGAGC .((((.((((((.......((((((((........))....((((((......))))))))))))(..(((........)))..)....))))))....))))....... ( -22.70) >consensus AGAUUAUUUUGUUUUCAAGGUAAUGGCACACACCAGCACACUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAAAGC .((((.((((((.......((((((((........))....((((((......))))))))))))((((((......))))))......))))))....))))....... (-26.55 = -26.67 + 0.11)

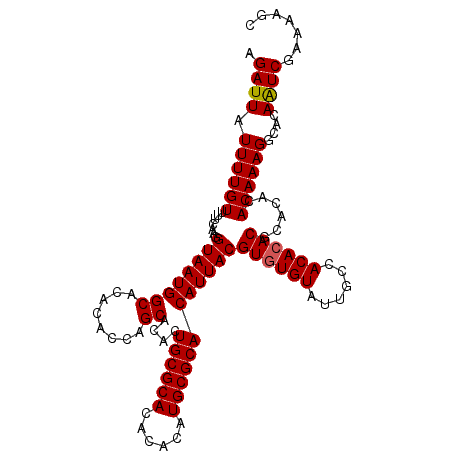

| Location | 17,971,268 – 17,971,378 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -34.17 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971268 110 - 22224390 GCUUUUCGACUGUGCCUUUGUGUACGUGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCU ..((((((((((((((..(((((((((..(((((......)))))...))))))))).....).)))))))(((.((((....)))).)))..))))))........... ( -36.80) >DroSec_CAF1 2006 110 - 1 GCUUUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCU .......((((((...(((((...((((((((((....)))))))))).((((((((....))))))))(.(((.((((....)))).))))......))))))))))). ( -35.60) >DroSim_CAF1 1369 110 - 1 GCUCUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACAUACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCU .......((((((...(((((..........((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))..........))))))))))). ( -33.95) >consensus GCUUUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAGUGUGCUGGUGUGUGCCAUUACCUUGAAAACAAAAUAAUCU .......((((((...(((((..........((((((.(((((..(((.((((((((....))))))))...)))..)))))))))))..........))))))))))). (-34.17 = -34.28 + 0.11)


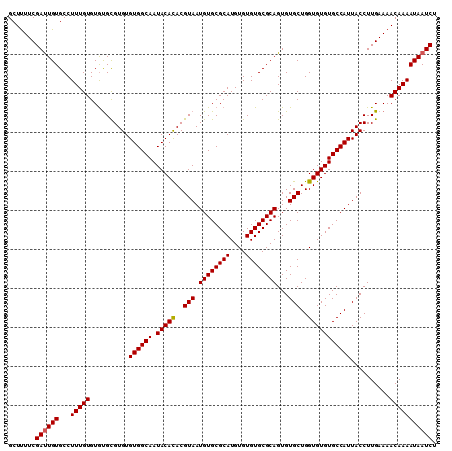
| Location | 17,971,308 – 17,971,417 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971308 109 + 22224390 CUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACACGUACACAAAGGCACAGUCGAAAAGCUUAUGACUUUUUGUUGUUUUUCUAUGCACUUUUCCAAAG .((((((......))))))...(((((((((........)))))))))...(((((((.((.((((((((....).)))))))........)).))).))))....... ( -28.50) >DroSec_CAF1 2046 109 + 1 CUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAAAGCUUAUGACUUUUUGUUGUUUUUCGAUGCAUUUUUUCAAAG .((((((......)))))).....(((((((......)))))))...........(((...(((((((((....(((.....)))))))))))))))............ ( -29.00) >DroSim_CAF1 1409 109 + 1 CUGCGCACACACAUGCGCACAUUACGUAUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAGAGCUUAUGACUUUUUGUUGUUUUUCUAUGCACUUUUUCAAAG .((((((......))))))......(((((....(((.................)))((((.((((((((....).))))))))))).....)))))............ ( -21.33) >consensus CUGCGCACACACAUGCGCACAUUACGUGUGUAUUGCCACACACGCACACACAAAGGCACAAUCGAAAAGCUUAUGACUUUUUGUUGUUUUUCUAUGCACUUUUUCAAAG .((((((......))))))......((((((......))))))(((.....(((((((....((((((((....).))))))).)))))))...)))............ (-22.57 = -22.80 + 0.23)

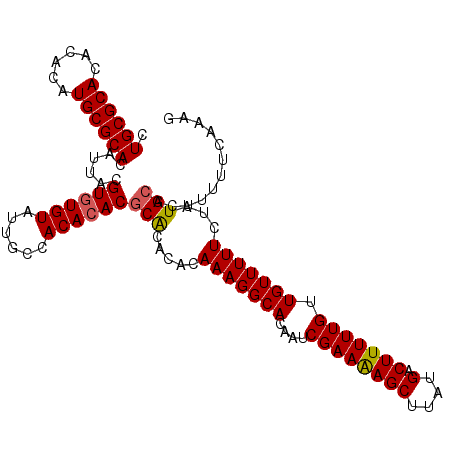

| Location | 17,971,308 – 17,971,417 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -32.00 |
| Energy contribution | -31.90 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17971308 109 - 22224390 CUUUGGAAAAGUGCAUAGAAAAACAACAAAAAGUCAUAAGCUUUUCGACUGUGCCUUUGUGUACGUGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAG ..........(((((((((...(((.(.((((((.....)))))).)..)))...)))))))))((((((((....))))))))....((((((((....)))))))). ( -35.80) >DroSec_CAF1 2046 109 - 1 CUUUGAAAAAAUGCAUCGAAAAACAACAAAAAGUCAUAAGCUUUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAG ..........(((((.((((..(((((.((((((.....)))))).).))))...)))))))))(((((((((....)))))))))..((((((((....)))))))). ( -33.40) >DroSim_CAF1 1409 109 - 1 CUUUGAAAAAGUGCAUAGAAAAACAACAAAAAGUCAUAAGCUCUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACAUACGUAAUGUGCGCAUGUGUGUGCGCAG ..........(((((((........(((.((((.(((((.........))))).)))).))).((((((((((....)))))))))).))))))).((((....)))). ( -31.20) >consensus CUUUGAAAAAGUGCAUAGAAAAACAACAAAAAGUCAUAAGCUUUUCGAUUGUGCCUUUGUGUGUGCGUGUGUGGCAAUACACACGUAAUGUGCGCAUGUGUGUGCGCAG ..........(((((((((...(((((.((((((.....)))))).).))))...)))))))))(((((((((....)))))))))..((((((((....)))))))). (-32.00 = -31.90 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:39 2006