
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,952,390 – 17,952,483 |
| Length | 93 |
| Max. P | 0.960422 |

| Location | 17,952,390 – 17,952,483 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -8.37 |
| Energy contribution | -8.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.73 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
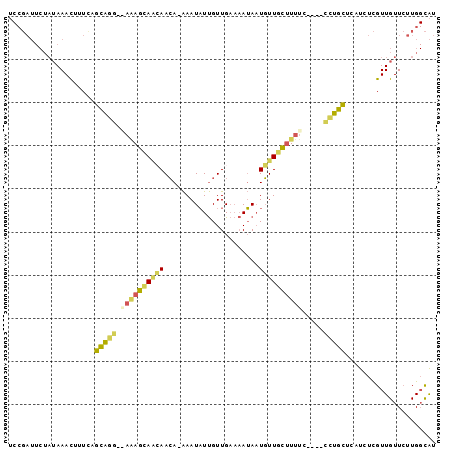
>X_DroMel_CAF1 17952390 93 + 22224390 UCCGAUUCUAUAAACUUUCAGCAGG-GAGAGCAACAACA-AAAUAUUGUUGAAAAUAAUGUUGCUUUUC----CCUGCUCAUCUCGUUGUUCUUGGCAU .((((....((((......((((((-(((((((((((((-(....)))).........)))))).))))----)))))).......))))..))))... ( -26.22) >DroSec_CAF1 11579 93 + 1 UCCGAUUCUAUAAACUUUCAGCAGG-GAGAGCAACAACA-AAAUAUUGUUGAAAAUAAUGUUGCUUUUC----CCUGCUCAUCUCGUUGUUCUUGGCAU .((((....((((......((((((-(((((((((((((-(....)))).........)))))).))))----)))))).......))))..))))... ( -26.22) >DroEre_CAF1 12279 96 + 1 UCCGAUUCUAUAAACUUUCAGCAGG--AAAGCAAAAACA-AAAUAUUGUUGAAAAUAAUGUUGCUUUUGCUAUUCUGCGCCUCUCGUUGUUCUUGGCAU ....................(((((--(.(((((((...-.((((((((.....))))))))..))))))).))))))(((.............))).. ( -23.92) >DroYak_CAF1 12288 92 + 1 UCCGAUUCUAUAAACUUUUUGCAGG--AAAGCAGCAACA-AAAUAUUGUUGAAAAUAAUGUUCCUUUUC----UUUGCUCUUCUCGUUGUUCUUGGCAU ...................((((((--(..(((((((((-(....)))))(((((.........)))))----............))))))))).))). ( -12.00) >DroMoj_CAF1 17351 80 + 1 -----------AAAUUUUGUGUUGAAA--AAUACAAAAACGAAUUUCGUUGCUCAAA-UUUUGUAUUUC----GAAAUUGUUAUUGUUGUUGCUG-UGU -----------....((((((((....--))))))))(((.(((((((.(((.....-....)))...)----))))))))).............-... ( -14.80) >consensus UCCGAUUCUAUAAACUUUCAGCAGG__AAAGCAACAACA_AAAUAUUGUUGAAAAUAAUGUUGCUUUUC____CCUGCUCAUCUCGUUGUUCUUGGCAU ....................(((((.((((((((((......................)))))))))).....)))))..................... ( -8.37 = -8.25 + -0.12)


| Location | 17,952,390 – 17,952,483 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -2.12 |
| Energy contribution | -3.40 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.12 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
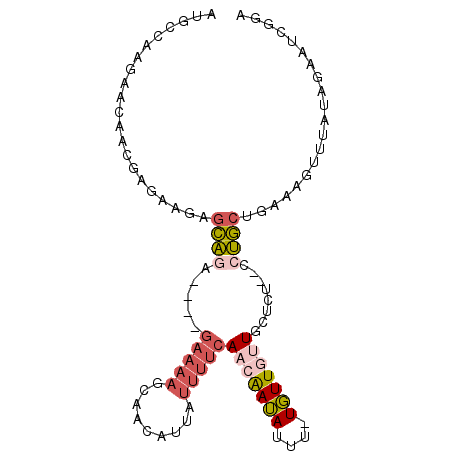
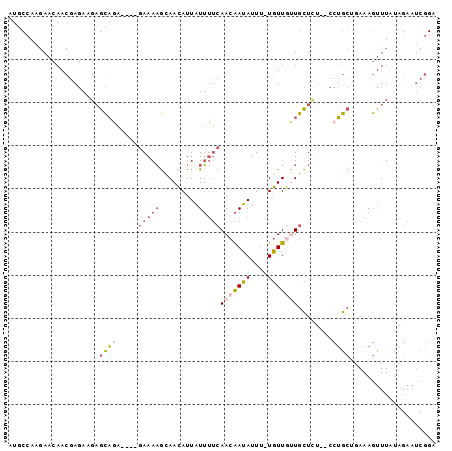
>X_DroMel_CAF1 17952390 93 - 22224390 AUGCCAAGAACAACGAGAUGAGCAGG----GAAAAGCAACAUUAUUUUCAACAAUAUUU-UGUUGUUGCUCUC-CCUGCUGAAAGUUUAUAGAAUCGGA ...((.....((......))((((((----((..(((((((.........((((....)-)))))))))).))-))))))................)). ( -23.80) >DroSec_CAF1 11579 93 - 1 AUGCCAAGAACAACGAGAUGAGCAGG----GAAAAGCAACAUUAUUUUCAACAAUAUUU-UGUUGUUGCUCUC-CCUGCUGAAAGUUUAUAGAAUCGGA ...((.....((......))((((((----((..(((((((.........((((....)-)))))))))).))-))))))................)). ( -23.80) >DroEre_CAF1 12279 96 - 1 AUGCCAAGAACAACGAGAGGCGCAGAAUAGCAAAAGCAACAUUAUUUUCAACAAUAUUU-UGUUUUUGCUUU--CCUGCUGAAAGUUUAUAGAAUCGGA ..(((.............)))((((...(((((((((((...((((......))))..)-))))))))))..--.)))).................... ( -21.62) >DroYak_CAF1 12288 92 - 1 AUGCCAAGAACAACGAGAAGAGCAAA----GAAAAGGAACAUUAUUUUCAACAAUAUUU-UGUUGCUGCUUU--CCUGCAAAAAGUUUAUAGAAUCGGA .............(((....(((...----(((((.........)))))(((((....)-)))))))((...--...))...............))).. ( -11.30) >DroMoj_CAF1 17351 80 - 1 ACA-CAGCAACAACAAUAACAAUUUC----GAAAUACAAAA-UUUGAGCAACGAAAUUCGUUUUUGUAUU--UUUCAACACAAAAUUU----------- ...-......................----(((((((((((-..((((........)))).)))))))))--))..............----------- ( -8.60) >consensus AUGCCAAGAACAACGAGAAGAGCAGA____GAAAAGCAACAUUAUUUUCAACAAUAUUU_UGUUGUUGCUCU__CCUGCUGAAAGUUUAUAGAAUCGGA .....................((((.....(((((.........)))))(((((((....)))))))........)))).................... ( -2.12 = -3.40 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:25 2006