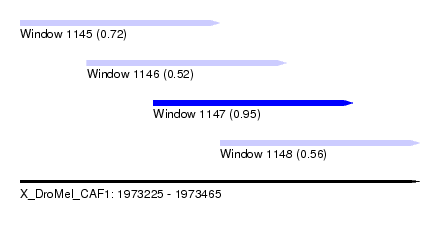
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,973,225 – 1,973,465 |
| Length | 240 |
| Max. P | 0.945926 |

| Location | 1,973,225 – 1,973,345 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -42.53 |
| Consensus MFE | -38.69 |
| Energy contribution | -39.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973225 120 + 22224390 UUCUUCCACACUCCAACUGCUGAUGGGAGGGCGAUCGGAUCCAUUUAUAUUCGAGGAUGGUAUUGCUAGGACCGGCGCAACCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGC ..(((((((......((((((..(((...(((..((((((........))))))(((((((..((((......))))..)))).))).)))....)))..))))))......))))))). ( -42.40) >DroSec_CAF1 6106 120 + 1 UUCUUCCACACUCCAACUGCUGAUGGGUGGGCGAUCGGAUCCAUUUACAUUCGAGGAUGGUAUUGCUAGAACCGGCGCAACCAUUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGC ..(((((((......((((((..(((.(.(((..((((((........))))))(((((((..((((......))))..)))))))..))).)..)))..))))))......))))))). ( -42.70) >DroYak_CAF1 6340 120 + 1 UUCUUCCACACUCCAACUGCUGAUGGGAGGGCGAUCGGUUCCAUUUACAUUGGAGGAUGGUAAUGCUAGAACCGGCGUAACCACUCCAGCUGAAACCACUGGCAAUGGCAAAGUGGAAGC ..(((((((...(((..((((..(((...(((.......((((.......))))(((((((.(((((......))))).)))).))).)))....)))..)))).)))....))))))). ( -42.50) >consensus UUCUUCCACACUCCAACUGCUGAUGGGAGGGCGAUCGGAUCCAUUUACAUUCGAGGAUGGUAUUGCUAGAACCGGCGCAACCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGC ..(((((((......((((((..(((...(((..((((((........))))))(((((((..((((......))))..)))).))).)))....)))..))))))......))))))). (-38.69 = -39.47 + 0.78)

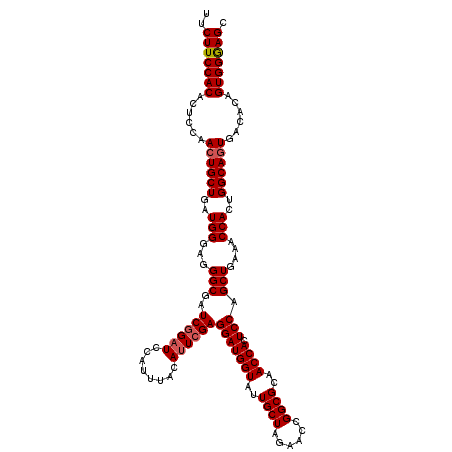
| Location | 1,973,265 – 1,973,385 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973265 120 + 22224390 CCAUUUAUAUUCGAGGAUGGUAUUGCUAGGACCGGCGCAACCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGU .........((((.(((((((..((((......))))..)))).)))((((....((((((.........)))))).)))).((((..((((((.......))))))..))))))))... ( -37.20) >DroSec_CAF1 6146 120 + 1 CCAUUUACAUUCGAGGAUGGUAUUGCUAGAACCGGCGCAACCAUUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGU ......((.((((.(((((((..((((......))))..))))))).((((....((((((.........)))))).)))).((((..((((((.......))))))..)))))))).)) ( -37.40) >DroYak_CAF1 6380 120 + 1 CCAUUUACAUUGGAGGAUGGUAAUGCUAGAACCGGCGUAACCACUCCAGCUGAAACCACUGGCAAUGGCAAAGUGGAAGCUAUUGCAAUCGGCAAUGGUAAUGUCAGGGGCAACGAAAGU ((.......((((((..((((.(((((......))))).))))))))))((((..(((((.((....))..)))))..((((((((.....))))))))....))))))...((....)) ( -41.30) >consensus CCAUUUACAUUCGAGGAUGGUAUUGCUAGAACCGGCGCAACCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGU ((............(((((((..((((......))))..)))).)))..((((..((((((.........))))))..(((.((((.....)))).)))....))))))...((....)) (-33.92 = -34.03 + 0.11)

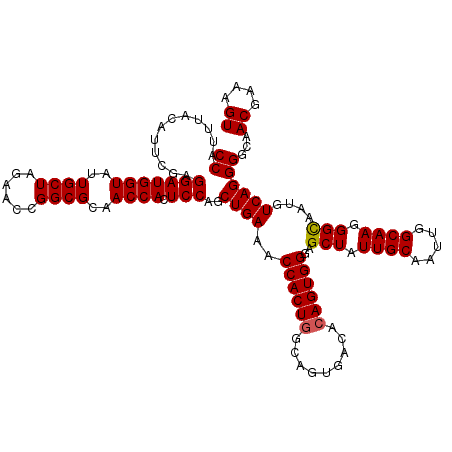
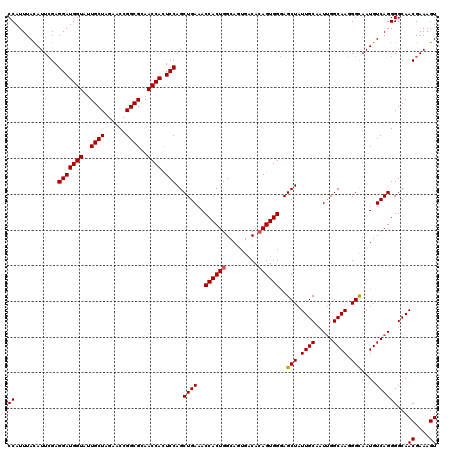
| Location | 1,973,305 – 1,973,425 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973305 120 + 22224390 CCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUCGACAUAGCCGCCAGCACUGGCAUCGAAA ....(((((((....((((((.........)))))).)))).((((.....)))))))(.(((((((.(((.((....))(((....(((....))).))))))....))))))).)... ( -39.40) >DroSec_CAF1 6186 120 + 1 CCAUUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUUGACAUAGCCGCCAGCACUGGCAUUGAAA ....(((((((....((((((.........)))))).)))).((((.....)))))))(((((((((.(((.((....))(((....(((....))).))))))....)))))))))... ( -43.60) >DroYak_CAF1 6420 120 + 1 CCACUCCAGCUGAAACCACUGGCAAUGGCAAAGUGGAAGCUAUUGCAAUCGGCAAUGGUAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUAGACAUAGCCGCUAGCACUGGCAUUGAAA .....((((((((..(((((.((....))..)))))..((....))..)))))..)))(((((((((.(....)(..((((((....(((....))).))))))..).)))))))))... ( -39.90) >consensus CCACUCCAGCUGAAACCACUGGCAGUGACACAGUGGGAGCUAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUAGACAUAGCCGCCAGCACUGGCAUUGAAA ........(((((..((((((.........))))))..((....))..))))).....(((((((((.(((.((....))(((....(((....))).))))))....)))))))))... (-38.29 = -38.30 + 0.01)

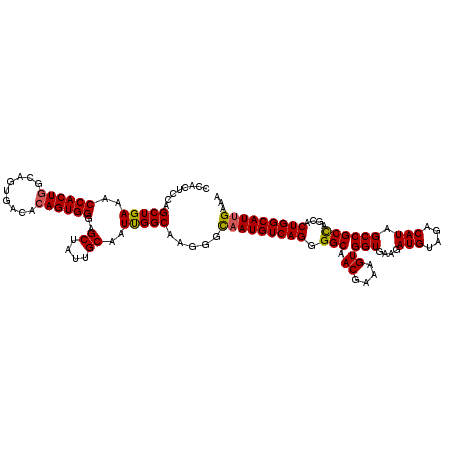
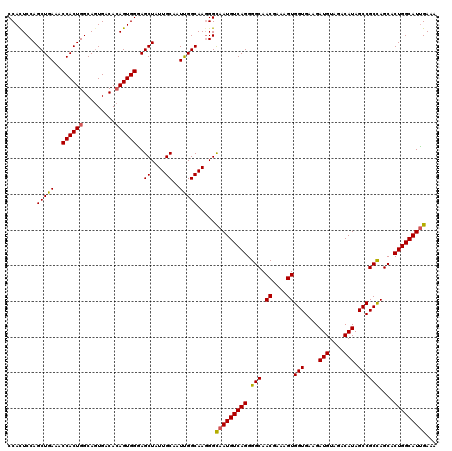
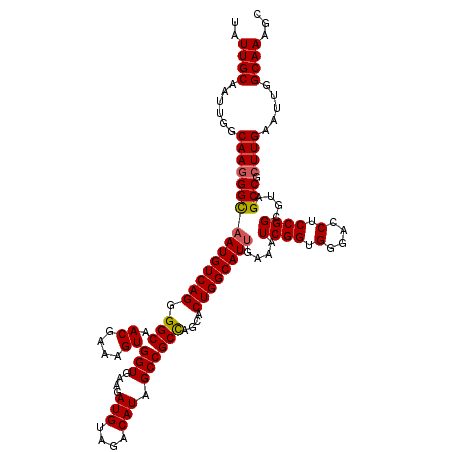
| Location | 1,973,345 – 1,973,465 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -37.81 |
| Energy contribution | -38.03 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1973345 120 + 22224390 UAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUCGACAUAGCCGCCAGCACUGGCAUCGAAAUCGGUGGGACCUCCGGCGUAGCCGCUUGAAUUGGCAAAGC ..((((......(((((((.(((((.(((((.((....))(((....(((....))).)))((((....))))((((....))))..).)))).))))).))).)))).....))))... ( -41.00) >DroSec_CAF1 6226 120 + 1 UAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUUGACAUAGCCGCCAGCACUGGCAUUGAAAUCGGUGGGACCUCCGGCGUAGCCGCUUGAAUUGGCAAAGC ..((((......(((((((((((((((.(((.((....))(((....(((....))).))))))....))))))))....((((.(....).))))....))).)))).....))))... ( -42.40) >DroYak_CAF1 6460 120 + 1 UAUUGCAAUCGGCAAUGGUAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUAGACAUAGCCGCUAGCACUGGCAUUGAAAUCGGUGAAACCUCCGGUGUAGCCGCUUGAAUUGGCAAUGC ((((((.....))))))((....((((((((......((((((....(((....))).)))))).((((((((((((....)))))......))))))).))).)))))....))..... ( -38.90) >consensus UAUUGCAAUUGGCAAGGGCAAUGUCAGGGGCAACGAAAGUGGUGAAGAUGUAGACAUAGCCGCCAGCACUGGCAUUGAAAUCGGUGGGACCUCCGGCGUAGCCGCUUGAAUUGGCAAAGC ..((((......(((((((((((((((.(((.((....))(((....(((....))).))))))....))))))))....((((.(....).))))....))).)))).....))))... (-37.81 = -38.03 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:52 2006