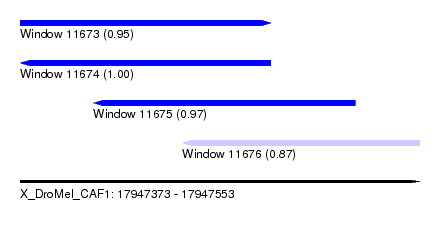
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,947,373 – 17,947,553 |
| Length | 180 |
| Max. P | 0.996729 |

| Location | 17,947,373 – 17,947,486 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -30.13 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17947373 113 + 22224390 -------GGAAACUUACCCUGAGUUUCUUCUUCCUCUCUGGGGUCAUGAAACCCUUCUUGGCCUUCUUGGCCUUGGAGGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG -------((((((((.....))))))))......((((..((((((.(((..((.....))..))).))))))..))))..........((((..(((..........)))..))))... ( -38.90) >DroPse_CAF1 10220 113 + 1 -------CAGCACUUACCCUGAGUUUCUUCUUCCUUUCUGGGGUCAUGAAACCCUUUUUGGCCUUCUUGGCUUUGGAAGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG -------.(((((((.....))))...........(((..((((((.(((..((.....))..))).))))))..))))))........((((..(((..........)))..))))... ( -30.50) >DroGri_CAF1 9988 120 + 1 UGGUUGCAGACACUUACCCGGAGUUUCUUCUUCCUCUCUGGGGUCAUGAAACCCUUCUUGGCCUUCUUGGCCUUAGAGGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG .((..(((((.((((.....)))).))).......(((((((((((.(((..((.....))..))).)))))))))))))..)).....((((..(((..........)))..))))... ( -40.10) >DroWil_CAF1 10178 113 + 1 -------GGAAACUUACCCUGAGUUUCUUCUUCCUUUCUGGUGUCAUGAAACCCUUCUUGGCCUUCUUGGCCUUGGAGGCUUCUUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG -------((((...........(((((.....((.....))......)))))(((((..((((.....))))..)))))....))))..((((..(((..........)))..))))... ( -36.40) >DroAna_CAF1 4334 113 + 1 -------CGUAACUCACCCUGAGUUUCUUCUUCCUCUCUGGGGUCAUGAAACCCUUCUUGGCCUUCUUGGCCUUGGAGGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG -------.(.(((((.....))))).).......((((..((((((.(((..((.....))..))).))))))..))))..........((((..(((..........)))..))))... ( -36.70) >DroPer_CAF1 8541 113 + 1 -------CAGCACUUACCCUGAGUUUCUUCUUCCUUUCUGGGGUCAUGAAACCCUUUUUGGCCUUCUUGGCUUUGGAAGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG -------.(((((((.....))))...........(((..((((((.(((..((.....))..))).))))))..))))))........((((..(((..........)))..))))... ( -30.50) >consensus _______CGAAACUUACCCUGAGUUUCUUCUUCCUCUCUGGGGUCAUGAAACCCUUCUUGGCCUUCUUGGCCUUGGAGGCUUCCUCCAUGCGCUUGCGCACCUCAGCACGCUUGCGCUCG ...........((((.....))))..........((((((((((((.(((..((.....))..))).))))))))))))..........((((..(((..........)))..))))... (-30.13 = -29.38 + -0.75)

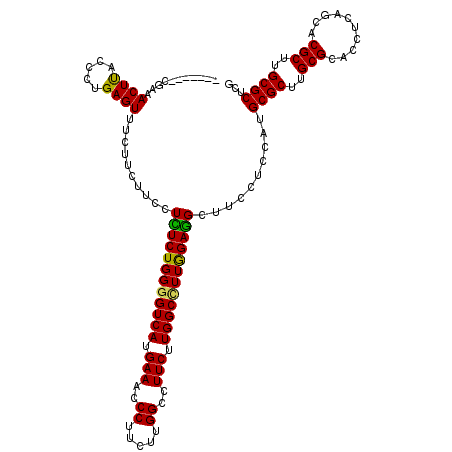

| Location | 17,947,373 – 17,947,486 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -36.17 |
| Energy contribution | -36.37 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17947373 113 - 22224390 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUUCC------- ...((((..(((..(....)..)))..)))).((((((...)))))).((((.....)))).....(((((....))))).............((((((.......)))))).------- ( -42.70) >DroPse_CAF1 10220 113 - 1 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCUUCCAAAGCCAAGAAGGCCAAAAAGGGUUUCAUGACCCCAGAAAGGAAGAAGAAACUCAGGGUAAGUGCUG------- ..(((((..((...(((((((((....))))..........(((((...(((.....)))......(((((....)))))......)))))......))))).))..))))).------- ( -39.50) >DroGri_CAF1 9988 120 - 1 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCUAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCCGGGUAAGUGUCUGCAACCA ...((((..(((..(....)..)))..)))).(((((....(((((..((((.....)))).....(((((....)))))...))))).........)))))(((...((....))))). ( -45.12) >DroWil_CAF1 10178 113 - 1 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAAGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACACCAGAAAGGAAGAAGAAACUCAGGGUAAGUUUCC------- ...((((..(((..(....)..)))..))))..(((((..((((....((((.....)))).....(((((((......((.....)).....))))))).))))...)))))------- ( -40.30) >DroAna_CAF1 4334 113 - 1 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUGAGUUACG------- ...((((..(((..(....)..)))..)))).((((((...)))))).((((.....)))).....(((((....))))).............(.(((((.....))))).).------- ( -43.50) >DroPer_CAF1 8541 113 - 1 CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCUUCCAAAGCCAAGAAGGCCAAAAAGGGUUUCAUGACCCCAGAAAGGAAGAAGAAACUCAGGGUAAGUGCUG------- ..(((((..((...(((((((((....))))..........(((((...(((.....)))......(((((....)))))......)))))......))))).))..))))).------- ( -39.50) >consensus CGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAAAGGAAGAAGAAACUCAGGGUAAGUGUCG_______ ...((((..(((..(....)..)))..)))).((((((...)))))).((((.....)))).....(((((((......((.....)).....))))))).................... (-36.17 = -36.37 + 0.20)



| Location | 17,947,406 – 17,947,524 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.43 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -37.83 |
| Energy contribution | -38.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17947406 118 - 22224390 --UUUAAUUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCC --...........((((.....)))).....((((((((((..((((..(((..(....)..)))..)))).((((((...)))))).((((.....))))......))))))).).)). ( -43.80) >DroVir_CAF1 10371 118 - 1 -CUGUG-CUUACAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGCGCCGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCC -.((((-(((....(((.....((((.....)))).......)))....((((((....)))))))))))))((((((...)))))).((((.....)))).....(((((....))))) ( -49.00) >DroGri_CAF1 10028 118 - 1 -CACUG-CUUACAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCUAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCC -.....-......((((.....((((....(((((....((.(((((..(((..(....)..)))..)))).).))...)))))....)))).....)))).....(((((....))))) ( -43.60) >DroSec_CAF1 6721 118 - 1 --UUUAAUUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGGGCAAGCGGGCUGAGAUGCCCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGCUUUCAUGACCC --.......(((..(((..((.((((.....))))....))...)))..((((((......))))..)))))((((((...)))))).((.((.(((((((.....))))))).)).)). ( -41.60) >DroWil_CAF1 10211 118 - 1 --AUUGCGUUCCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAAGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACAC --......(((((((((.....((((.....)))).)).))..((((..(((..(....)..)))..)))).)))))(((((((....((((.....)))).....)))))))....... ( -41.40) >DroMoj_CAF1 10364 119 - 1 CUUCUG-UUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGCGCCGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGCUUCAUGACCC ....((-(((...((((.....)))))))))((((........((((..((((((....))))))..)))).((((((...)))))).))))..((((.((.....)).))))....... ( -48.30) >consensus __UUUG_UUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCC .............((((.....((((....(((((....((.(((((..((((((....))))))..)))).).))...)))))....)))).....)))).....(((((....))))) (-37.83 = -38.83 + 1.00)

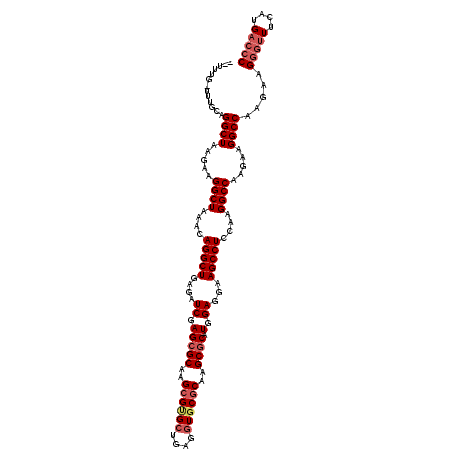

| Location | 17,947,446 – 17,947,553 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17947446 107 - 22224390 -----ACCAUGCUGAUACCCGGAUUGUUCGUU---CA--UUUAAUUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAA -----.((((((....((((((..((((((((---((--(((..((((..((((.....))))...))))...)))).)))))..))))..))).)))((....))))))))..... ( -33.10) >DroPse_CAF1 10293 100 - 1 -----ACGAUGGU-------CGAUUGUUUUUGG--UG--UUUGA-UUGCAGGCAAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAA -----.(((((((-------(....((((((..--((--((((.-...))))))..))))))......))))...)))).((((..(((..(....)..)))..))))......... ( -33.20) >DroWil_CAF1 10251 107 - 1 -------CAUCCUACUAAU-CGAUUCCAUUUGAAUCU--AUUGCGUUCCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAAGAA -------............-...((((((((((.(((--...((.(....((((.....))))....).))..)))))))((((..(((..(....)..)))..))))))))))... ( -31.80) >DroMoj_CAF1 10404 112 - 1 CGCCUACGAUGCUAAUUG-AGAUUUGUGUGUU---CGCUUCUG-UUUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGCGCCGAGGUGCGCAAGCGCAUGGAGGAA ..(((.(.((((......-.......((((((---((.(((.(-((((..((((.....))))...))))).)))..)))))))).((((((....))))))....)))).)))).. ( -41.30) >DroAna_CAF1 4407 91 - 1 --------UUGCUGAUC-------------UC---AA--UUUGAAUUUCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAA --------....(((((-------------((---(.--((((.......((((.....))))...)))).)))))))).((((..(((..(....)..)))..))))......... ( -29.80) >DroPer_CAF1 8614 100 - 1 -----ACGAUGGU-------CGAUUGUUUUUGG--UG--UUUGA-UUGCAGGCAAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAA -----.(((((((-------(....((((((..--((--((((.-...))))))..))))))......))))...)))).((((..(((..(....)..)))..))))......... ( -33.20) >consensus _____ACGAUGCU_AU____CGAUUGUUUUUG___CG__UUUGA_UUGCAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAA ...........................................................((((.....))))....(((.((((..((((((....))))))..)))).)))..... (-20.75 = -20.62 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:20 2006