
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,942,995 – 17,943,104 |
| Length | 109 |
| Max. P | 0.997614 |

| Location | 17,942,995 – 17,943,104 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -10.81 |
| Energy contribution | -14.01 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
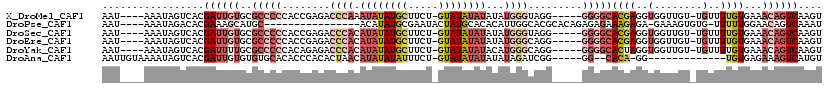
>X_DroMel_CAF1 17942995 109 + 22224390 AAU----AAAUAGUCACGAUUGUGCGCCCCCACCGAGACCCAAAUAUAUGCUUCU-GUAUAUAUAUAUGGGUAGG-----GGGGCACGAGGUGGUUGU-UGUUUUGUGAAACAGUCAAGU ...----.((((..(((..((((((.(((((......(((((.((((((((....-))))))))...))))).))-----))))))))).)))..)))-)((((....))))........ ( -40.60) >DroPse_CAF1 4984 98 + 1 AAU----AAAUAGACACGAAAGCAUGC----------------ACAUAUGCGAAUACUAUGCACACAUUGGCACGCACAGAGAGAAAGAGA-GAAAGUGUG-UUUUUGGAACAGUCAAAU ...----.....(((......(((((.----------------...)))))..............((..(((((((...............-....)))))-))..)).....))).... ( -16.41) >DroSec_CAF1 3134 109 + 1 AAU----AAAUAGUCACGAUUGUGCGCCCCCACCGAGACCCACAUAUAUGCUUCU-GUAUAUAUAUAUGGGUAGG-----GGGGCACGAGGUGGUUGU-UGUUUUGUGAAACAGUCAAGU ...----.((((..(((..((((((.(((((......(((((.((((((((....-))))))))...))))).))-----))))))))).)))..)))-)((((....))))........ ( -40.60) >DroEre_CAF1 3092 109 + 1 AAU----AAAUAGUCACGAUUGUGCGCCCCCACCGAGACCCACAUAUAUGCUUCU-GUAUAUAUAUAUGGGCAGG-----GGGGCACGAGGUGGUUGU-UGUUUUGUGAAACAGUCAAGU ...----.((((..(((..((((((.(((((.....(.((((.((((((((....-))))))))...))))).))-----))))))))).)))..)))-)((((....))))........ ( -38.30) >DroYak_CAF1 3071 109 + 1 AAU----AAAUAGUCACGAUUUUGCGCCCCCACAGAGACCCACAUAUAUGCUUCU-GUAUAUAUACAUGGGCAGG-----GGGGCACUAGGUGGUUGU-UGUUUUGUGAAACAGUCAAGU ...----((((((..((.((((.(.((((((.....(.((((.((((((((....-))))))))...)))))..)-----))))).).)))).))..)-)))))................ ( -32.30) >DroAna_CAF1 3034 98 + 1 AAUUGUAAAAUAGUCACGAUUGUGUGUGCACACCCACACUAACAUAUAUAUUUCU-GUAUAUAUAUAUAGAUCGG-----GG--CACA-GG-------------UGUGAGAAAGUCAUGU .............(((((.(((((((....).(((...(((..((((((((....-))))))))...)))...))-----))--))))-).-------------)))))........... ( -20.30) >consensus AAU____AAAUAGUCACGAUUGUGCGCCCCCACCGAGACCCACAUAUAUGCUUCU_GUAUAUAUAUAUGGGCAGG_____GGGGCACGAGGUGGUUGU_UGUUUUGUGAAACAGUCAAGU .................((((((..(((((........((((.((((((((.....))))))))...)))).........)))))((((..(........)..))))...)))))).... (-10.81 = -14.01 + 3.20)

| Location | 17,942,995 – 17,943,104 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -15.13 |
| Energy contribution | -16.99 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
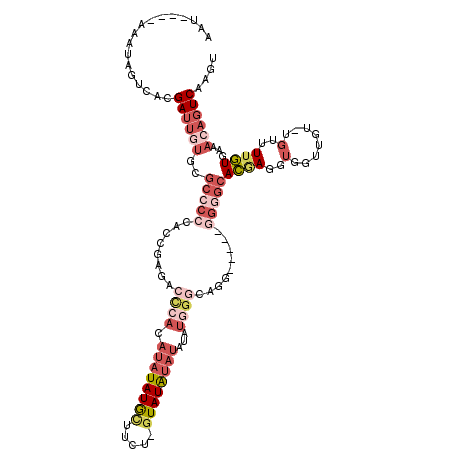
>X_DroMel_CAF1 17942995 109 - 22224390 ACUUGACUGUUUCACAAAACA-ACAACCACCUCGUGCCCC-----CCUACCCAUAUAUAUAUAC-AGAAGCAUAUAUUUGGGUCUCGGUGGGGGCGCACAAUCGUGACUAUUU----AUU ..(((..(((((....)))))-...........(((((((-----((((((((.((((((....-......)))))).)))))...)).)))))))).)))............----... ( -30.30) >DroPse_CAF1 4984 98 - 1 AUUUGACUGUUCCAAAAA-CACACUUUC-UCUCUUUCUCUCUGUGCGUGCCAAUGUGUGCAUAGUAUUCGCAUAUGU----------------GCAUGCUUUCGUGUCUAUUU----AUU .((((.......)))).(-(((......-...........(((((((..(....)..))))))).....((((....----------------..))))....))))......----... ( -17.70) >DroSec_CAF1 3134 109 - 1 ACUUGACUGUUUCACAAAACA-ACAACCACCUCGUGCCCC-----CCUACCCAUAUAUAUAUAC-AGAAGCAUAUAUGUGGGUCUCGGUGGGGGCGCACAAUCGUGACUAUUU----AUU ..(((..(((((....)))))-...........(((((((-----(((((((((((((((....-......))))))))))))...)).)))))))).)))............----... ( -34.20) >DroEre_CAF1 3092 109 - 1 ACUUGACUGUUUCACAAAACA-ACAACCACCUCGUGCCCC-----CCUGCCCAUAUAUAUAUAC-AGAAGCAUAUAUGUGGGUCUCGGUGGGGGCGCACAAUCGUGACUAUUU----AUU ..(((..(((((....)))))-...........(((((((-----(((((((((((((((....-......))))))))))))...)).)))))))).)))............----... ( -34.50) >DroYak_CAF1 3071 109 - 1 ACUUGACUGUUUCACAAAACA-ACAACCACCUAGUGCCCC-----CCUGCCCAUGUAUAUAUAC-AGAAGCAUAUAUGUGGGUCUCUGUGGGGGCGCAAAAUCGUGACUAUUU----AUU .......(((((....)))))-...........(((((((-----((.((((((((((((....-......))))))))))))....).))))))))................----... ( -32.00) >DroAna_CAF1 3034 98 - 1 ACAUGACUUUCUCACA-------------CC-UGUG--CC-----CCGAUCUAUAUAUAUAUAC-AGAAAUAUAUAUGUUAGUGUGGGUGUGCACACACAAUCGUGACUAUUUUACAAUU .(((((......((((-------------((-((..--(.-----.......((((((((((..-....))))))))))..)..)))))))).........))))).............. ( -21.26) >consensus ACUUGACUGUUUCACAAAACA_ACAACCACCUCGUGCCCC_____CCUACCCAUAUAUAUAUAC_AGAAGCAUAUAUGUGGGUCUCGGUGGGGGCGCACAAUCGUGACUAUUU____AUU .................................(((((((........((((((((((((...........)))))))))))).......)))))))....................... (-15.13 = -16.99 + 1.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:16 2006