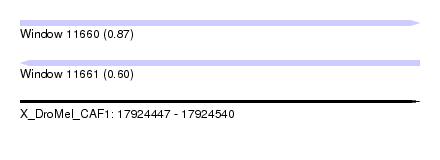
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,924,447 – 17,924,540 |
| Length | 93 |
| Max. P | 0.867074 |

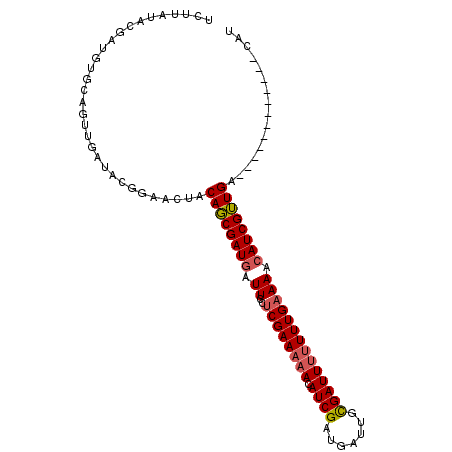
| Location | 17,924,447 – 17,924,540 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17924447 93 + 22224390 AUGACAUCAUCGAUAUCAUCGAUCUUUUCAAA-CAAUCACAAGCAUCGAUGUUUUUCGAGGAAUCAUCGAUGUUCUUCCAU---------ACAUCGGAGGACA .(((.(..((((((...))))))..).)))..-.........(((((((((..(((....))).)))))))))((((((..---------.....)))))).. ( -23.80) >DroSec_CAF1 1496 91 + 1 AUG------------UCAACGAUGUUUUCAAAAAAAUCGCAAUCAUCGAUGUUUUUCGAGGAAUCAUCGCUGUAGUUCCGUAUCAACUGCACAUCGUAUAAGA ...------------...((((((((((((((((.((((.......)))).))))).)))))........(((((((.......)))))))))))))...... ( -18.30) >DroSim_CAF1 400 91 + 1 AUG------------UCAACGAUGUUUACAAAAAAAUCGCAAUCAUCGAUGUUUUUCGAGGAAUCAUCGCUGUAGUUCUGUAUCAACUGCACAUCGUAUAAGA ...------------...(((((((.................((.((((......)))).)).........((((((.......)))))))))))))...... ( -17.60) >consensus AUG____________UCAACGAUGUUUUCAAAAAAAUCGCAAUCAUCGAUGUUUUUCGAGGAAUCAUCGCUGUAGUUCCGUAUCAACUGCACAUCGUAUAAGA ..................(((((((....................((((......))))((((..((....))..))))...........)))))))...... (-12.70 = -12.37 + -0.33)

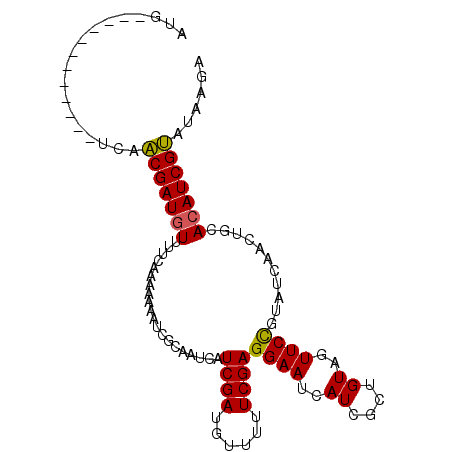

| Location | 17,924,447 – 17,924,540 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17924447 93 - 22224390 UGUCCUCCGAUGU---------AUGGAAGAACAUCGAUGAUUCCUCGAAAAACAUCGAUGCUUGUGAUUG-UUUGAAAAGAUCGAUGAUAUCGAUGAUGUCAU .......((((((---------........))))))(((((((.((((....(((((((.(((.......-......))))))))))...)))).)).))))) ( -23.02) >DroSec_CAF1 1496 91 - 1 UCUUAUACGAUGUGCAGUUGAUACGGAACUACAGCGAUGAUUCCUCGAAAAACAUCGAUGAUUGCGAUUUUUUUGAAAACAUCGUUGA------------CAU ((((((.((((.....))))))).)))....((((((((.((..((((((((.((((.......)))))))))))).)))))))))).------------... ( -19.90) >DroSim_CAF1 400 91 - 1 UCUUAUACGAUGUGCAGUUGAUACAGAACUACAGCGAUGAUUCCUCGAAAAACAUCGAUGAUUGCGAUUUUUUUGUAAACAUCGUUGA------------CAU ((((((.((((.....))))))).)))....((((((((..((.((((......)))).))((((((.....)))))).)))))))).------------... ( -20.90) >consensus UCUUAUACGAUGUGCAGUUGAUACGGAACUACAGCGAUGAUUCCUCGAAAAACAUCGAUGAUUGCGAUUUUUUUGAAAACAUCGUUGA____________CAU ...............................((((((((.((..((((((((.((((.......)))))))))))))).))))))))................ (-14.10 = -14.77 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:07 2006