
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,905,144 – 17,905,241 |
| Length | 97 |
| Max. P | 0.611110 |

| Location | 17,905,144 – 17,905,241 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.90 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -14.44 |
| Energy contribution | -15.50 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
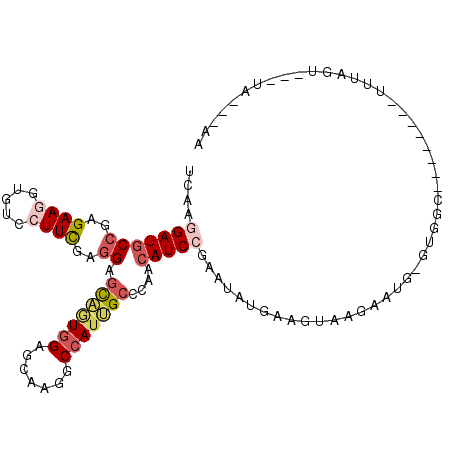
>X_DroMel_CAF1 17905144 97 + 22224390 UCAAGGAUGCCGAGAAGGUCUCCUUUGAGGAGCAGUGGAGCAAGGCCAUUGCCCAGCAUCCGAAUAUGACGUAAGAAUG-UUUGC--------UGUAGU---UACCAAA (((((((.(((.....))).))).))))((.(((((((.......))))))))).(((..(((((((..(....).)))-)))).--------)))...---....... ( -29.70) >DroVir_CAF1 6168 89 + 1 UAAAAGAUGCCGAGAAAUUGUCCUUCGAGGACUUUUGGGGCAGGGCCAUGGCCGUUCAUCCGAAUAUGGAGUGAGUAUCAGAAAAAA--------------------AA .......((((..((((..((((.....))))))))..)))).(((....))).(((((((......))).))))............--------------------.. ( -22.30) >DroSec_CAF1 5442 97 + 1 UCAAGGAUGCCGAGAAGGUGUCCUUCGAGGAGCAGUGGAGCAAGGCCAUUGCCCAACAUCCGAAUAUGACGUAAGAAUG-GUGGC--------UGUAGU---UACCAAA (((((((((((.....))))))))).))((.(((((((.......)))))))))...............(....)..((-(((((--------....))---))))).. ( -34.70) >DroEre_CAF1 5447 93 + 1 UCAAGGAUGCCGAGAAGAUGUCCUUCGAGGAGCAGUGGAGCAAGGCGAUUGCCCAACAUCCAAAUAUGAUGUGAGAAUG-UUGGC--------UUUAAU---UA----U ((((((((..(.....)..)))))).))(((((..(((.((((.....)))))))(((((.......))))).......-...))--------)))...---..----. ( -22.80) >DroWil_CAF1 5543 105 + 1 UAAAGGAUCCAGAGAAAAUCUCAUUCGAAGAGCAAUGGAGUCGUGCCAUGGCUUUGCAUCCCAAUAUGGAGUAAGU-UG-GCGGU--UUUUUCUUUUUUGAUUUCAAAA ..........((((((((((((((((.....).)))))......((((..((((.((.(((......)))))))))-))-)))))--))))))).(((((....))))) ( -22.30) >DroYak_CAF1 5452 93 + 1 UCAAAGAUGCCGAGAAGGUGUCCUUCGAGGAGCAGUGGAGUAAGGCCAUUGCCCUACAUCCGAAUAUGAGGUAAGAAUG-GUGGC--------CUUACU---UA----U .((..(...((..(((((...)))))..))..)..))((((((((((((((..((....((........))..))..))-)))))--------))))))---).----. ( -31.20) >consensus UCAAGGAUGCCGAGAAGGUGUCCUUCGAGGAGCAGUGGAGCAAGGCCAUUGCCCAACAUCCGAAUAUGAAGUAAGAAUG_GUGGC________UUUAGU___UA___AA ....(((((((..((((.....))))..)).(((((((.......)))))))....)))))................................................ (-14.44 = -15.50 + 1.06)


| Location | 17,905,144 – 17,905,241 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.90 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17905144 97 - 22224390 UUUGGUA---ACUACA--------GCAAA-CAUUCUUACGUCAUAUUCGGAUGCUGGGCAAUGGCCUUGCUCCACUGCUCCUCAAAGGAGACCUUCUCGGCAUCCUUGA .......---......--------.....-..................((((((((((((.(((.......))).))((((.....)))).....)))))))))).... ( -23.00) >DroVir_CAF1 6168 89 - 1 UU--------------------UUUUUUCUGAUACUCACUCCAUAUUCGGAUGAACGGCCAUGGCCCUGCCCCAAAAGUCCUCGAAGGACAAUUUCUCGGCAUCUUUUA ..--------------------.............(((.(((......))))))..(((....))).((((......((((.....))))........))))....... ( -18.24) >DroSec_CAF1 5442 97 - 1 UUUGGUA---ACUACA--------GCCAC-CAUUCUUACGUCAUAUUCGGAUGUUGGGCAAUGGCCUUGCUCCACUGCUCCUCGAAGGACACCUUCUCGGCAUCCUUGA ..((((.---......--------)))).-..................((((((((((((.(((.......))).))))....(((((...))))).)))))))).... ( -24.70) >DroEre_CAF1 5447 93 - 1 A----UA---AUUAAA--------GCCAA-CAUUCUCACAUCAUAUUUGGAUGUUGGGCAAUCGCCUUGCUCCACUGCUCCUCGAAGGACAUCUUCUCGGCAUCCUUGA .----..---......--------.((((-(((((.............)))))))))......(((..((......)).....((((.....))))..)))........ ( -20.52) >DroWil_CAF1 5543 105 - 1 UUUUGAAAUCAAAAAAGAAAAA--ACCGC-CA-ACUUACUCCAUAUUGGGAUGCAAAGCCAUGGCACGACUCCAUUGCUCUUCGAAUGAGAUUUUCUCUGGAUCCUUUA (((((....)))))........--.....-..-..............(((((.((.(((.((((.......)))).)))........(((.....))))).)))))... ( -16.80) >DroYak_CAF1 5452 93 - 1 A----UA---AGUAAG--------GCCAC-CAUUCUUACCUCAUAUUCGGAUGUAGGGCAAUGGCCUUACUCCACUGCUCCUCGAAGGACACCUUCUCGGCAUCUUUGA .----..---((((((--------((((.-...((((((.((.......)).))))))...))))))))))....((((....(((((...)))))..))))....... ( -28.10) >consensus UU___UA___ACUAAA________GCCAC_CAUUCUUACGUCAUAUUCGGAUGUAGGGCAAUGGCCUUGCUCCACUGCUCCUCGAAGGACACCUUCUCGGCAUCCUUGA ................................................(((((((((((....))).................((((.....)))).)))))))).... (-12.86 = -13.70 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:58 2006