
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,902,203 – 17,902,311 |
| Length | 108 |
| Max. P | 0.698887 |

| Location | 17,902,203 – 17,902,311 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.83 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -25.23 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17902203 108 + 22224390 G------------AAACAGAAAGCUGGAUGGACUGCCAGGGAGCUUGGAUCUACCUGGAGGCCAUCUUUGCCUCGGCGGAUAUUCAGCGACAGCUGCCCCACGAGGCCAAGAUGUUCUUC .------------............((.((((...(((((...))))).)))))).((((..((((((.((((((..((.....((((....))))..)).)))))).))))))..)))) ( -39.20) >DroPse_CAF1 2557 120 + 1 CUCUCUCUCUCUCCUCUAGAGGCCUGGAUGGACUGUCAGGGUGCAUGGAUCUACCUGGAGGCUAUUUUCGCCUCGGCGGAUAUUCAGCGCCAGUUGCCCCAAGAGGCCAAGAUGUUCUUU .......(((..(((((.(.(((((((.(((((((((.(((((........))))).(((((.......))))))))))...))))...))))..))).).)))))...)))........ ( -38.60) >DroGri_CAF1 3067 104 + 1 ----------------CAGAGGCUUGGUUGGACUGCCAGGGCUCUUGGGUCUACUUGGAGGCAAUCUUUGCCUCGGCCGACAUACAGCGCCAGCUGCCCAAUGAGGCCAAGAUGUUCAAC ----------------....(((((.(((((.(.(((.(((((....))))).....((((((.....))))))))).).....((((....)))).))))).)))))............ ( -41.10) >DroWil_CAF1 2542 109 + 1 U-----------UUUUCAGAGGCUUGGAUGGAUUGCCAAGGUGCCUGGAUCUAUUUGGAGGCAAUUUUCGCCUCGGCUGAUAUACAACGUCAAUUGCCAAAUGAGGCGAAAAUGUUUUUU .-----------..(((((.(.(((((........))))).)..))))).......(((((((.(((((((((((((((((.......))))...)))....))))))))))))))))). ( -34.00) >DroMoj_CAF1 4051 112 + 1 --------UCGGACCUUAGAGGCUUGGAUGGAUUGCCAGGGCGCCUGGAUCUAUUUGGAAGCCAUCUUUGCCUCGGCUGACAUACAACGGCAGCUGCCCCACGAAGCCAAAAUGUUCAAU --------..((((....(((((..((((((....(((((...))))).(((....)))..))))))..)))))((((..........(((....)))......)))).....))))... ( -39.19) >DroPer_CAF1 2563 120 + 1 CUCUCUCUCUCUCCUCUAGAGGCCUGGAUGGACUGUCAGGGUGCAUGGAUCUACUUGGAGGCUAUCUUCGCCUCGGCGGAUAUUCAGCGCCAGUUGCCCAACGAGGCCAAGAUGUUCUUU .......(((..((((....((((((((((..(((((..((((.((((.(((....)))..))))...))))..))))).))))))).))).(((....)))))))...)))........ ( -37.20) >consensus _____________CUCCAGAGGCCUGGAUGGACUGCCAGGGUGCAUGGAUCUACUUGGAGGCAAUCUUCGCCUCGGCGGAUAUACAGCGCCAGCUGCCCAACGAGGCCAAGAUGUUCUUU ..................(((((..((((((.((.(((((.............))))).))))))))..)))))(((.......((((....)))).........)))............ (-25.23 = -24.96 + -0.27)

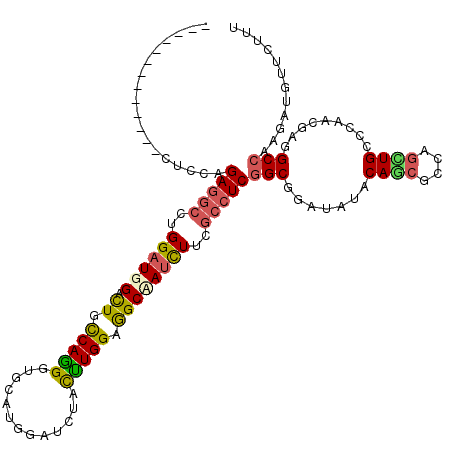
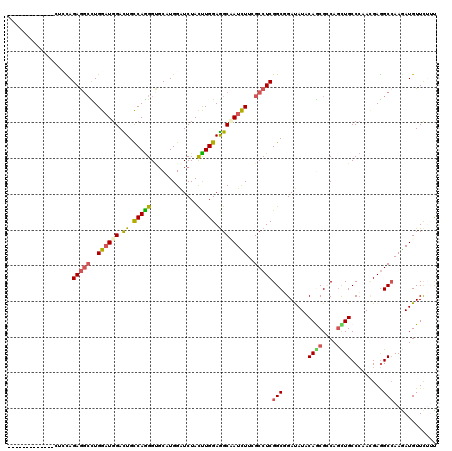
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:56 2006